Merow, C., Silander, J. A. (2014), A comparison of Maxlike and Maxent for modelling species distributions. Methods in Ecology and Evolution, 5: 215–225. doi: 10.1111/2041-210X.12152
—
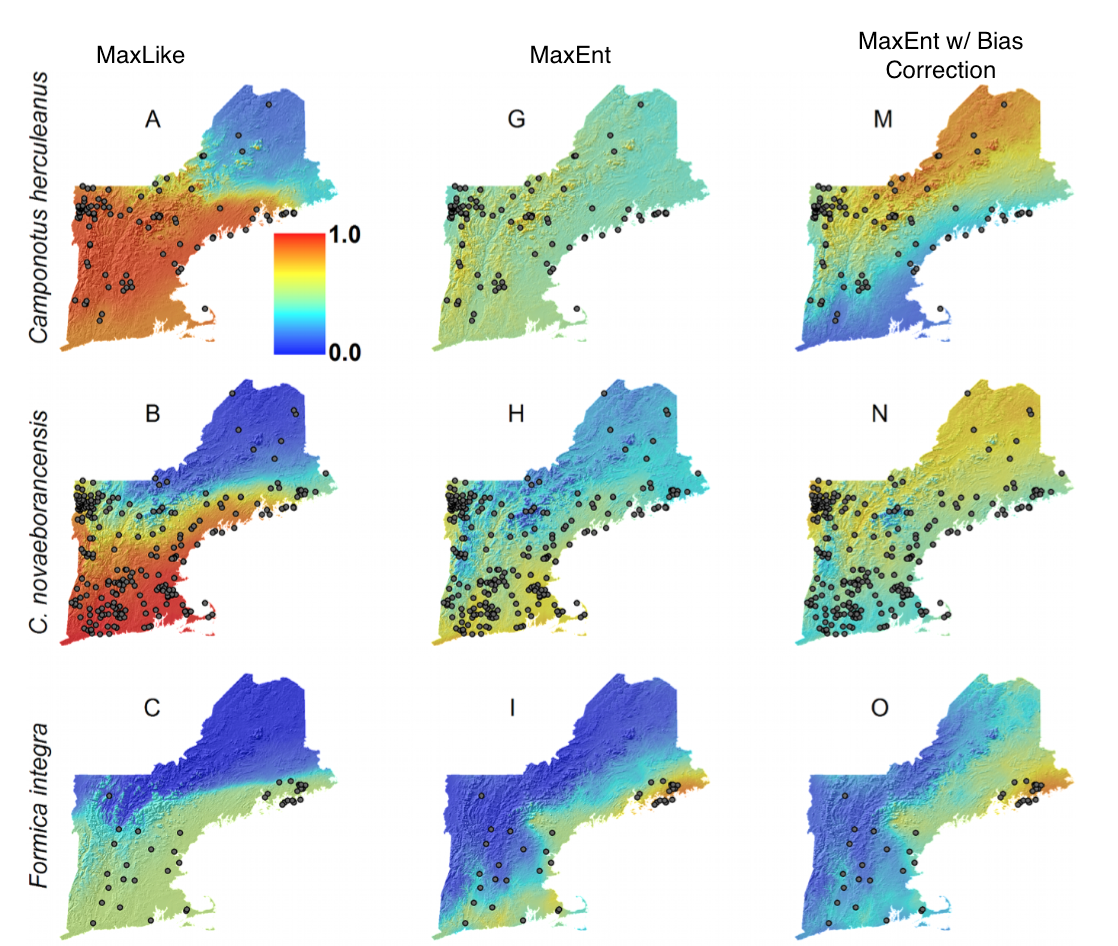
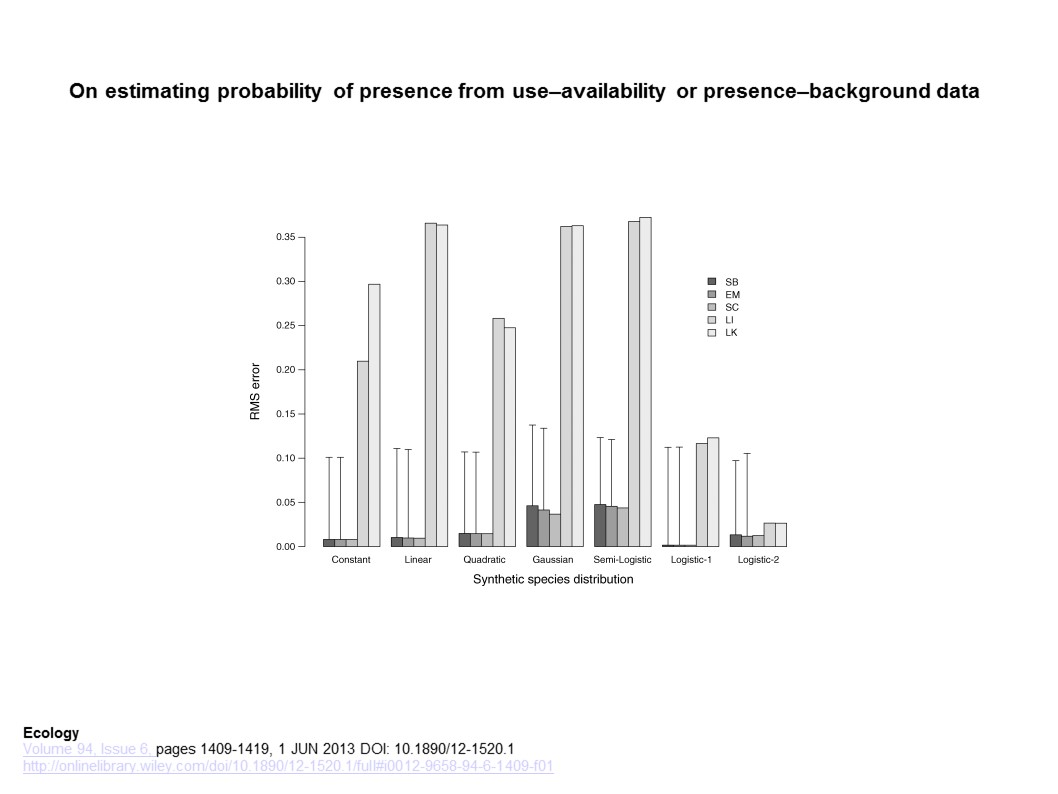
Here, the authors compare MaxLike (presence only method set up by Royle et al. 2012) to MaxEnt (widely used presence-background method). They detail how MaxEnt and MaxLike compare in their structure, providing instances of when predictions between the two would differ, and then linking the two through a discussion of sampling assumptions. The authors advocate for the use of MaxEnt’s raw output (relative occurrence rate), and point out reasons that the raw output is not equivalent to an occurrence probability (e.g., λ(x) may be larger than 1).
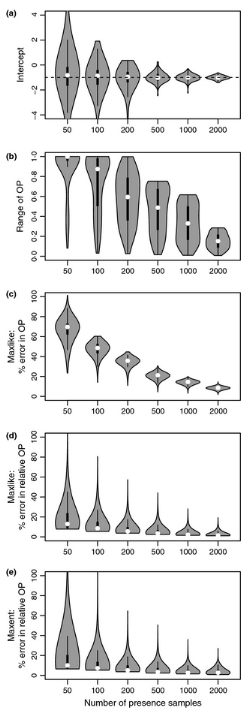
The authors further show the sensitivity of MaxLike to smaller sample sizes (Figure 3), using both empirical (Carolina wren data), and simulated data (using same model as Royle et al. 2012). MaxLike and MaxEnt outputs were strikingly similar (correlation of 0.999) when considering the raw output (relative occurrence rate) as the unit being compared between approaches. In some circumstances (low sample size), it may be difficult for Maxlike to estimate the intercept (β0) value.
In the end, the authors offer a defense of MaxEnt, and argue that both MaxLike and MaxEnt may make strong assumptions. For instance, MaxEnt assumes that the data are a random sample of individuals (though don’t both methods make this assumption?), and makes the assumption that the loglinear model is appropriate for the count data (which is defensible). Basically, if sample size is large and detection probability is constant, Maxlike is preferred since it can directly estimate occurrence probability. If sample size is small, and the model is more focused on habitat suitability instead of actual occurrence probability, the raw output (relative occurrence rate) of MaxEnt may be preferred.