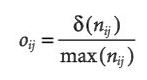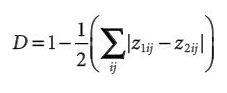Fourcade, Y., et al. (2014). “Mapping species distributions with MAXENT using a geographically biased sample of presence data: a performance assessment of methods for correcting sampling bias.” PLoS One 9(5): e97122.
http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0097122
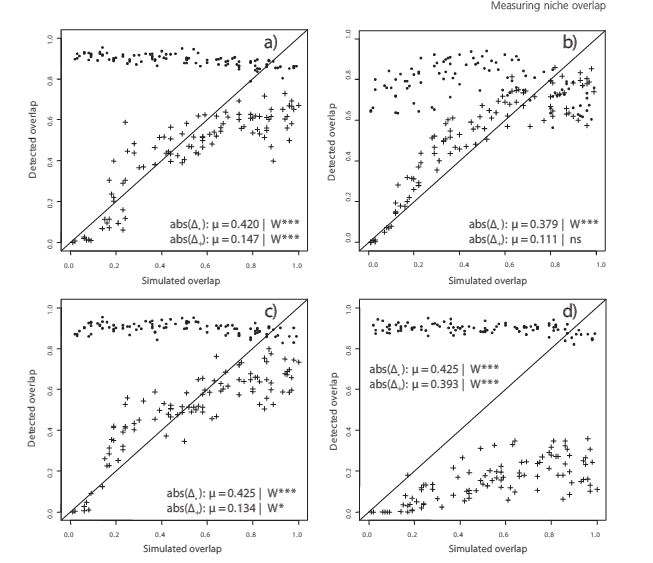
Fourcade et al. attempt to assess the effectiveness of a number of methods for correcting sampling bias in species distribution modeling with MaxEnt. Bias in species sampling has been well established as an important and difficult problem in species distribution modeling. The authors take large, and likely spatially unbiased, presence only sample sets for 1 virtual and 2 real species and impose 4 types of spatial bias on them to simulate sampling bias. These types of biases are (1) Two Areas, the northern region has high sample density and the southern region low density, (2) Gradient, a density gradient decreasing from north to south, (3) Center, the density decreases gradually from the core of the distribution to the edges, (4) Travel Time, probability of keeping a record was highest when it had the lowest travel time to the nearest city. 5 different data-processing methods were used to limit the effect of these spatial biases: (a) Systematic Sampling, a grid of a defined cell size was superimposed on the distribution and 1 record was chosen per grid cell, (b) Bias File, MaxEnt can be given a file representing sampling effort with which it weights the sampled points, (c) Restricted Background, MaxEnt’s background points were drawn exclusively from buffer areas around biased occurrences, (d) Cluster, a PCA was performed on the environmental predictors then occurrence points were analyzed for clustering in the 2 dimensional environmental PCA space and 1 record was randomly sampled per cluster, (e) Split, occurrences were split into a northern and southern group and MaxEnt was applied independently to each area. These methods all seem relatively well grounded individually but the combination fails to make much sense. Most notably, it is entirely unclear why grid based selection is used for spatial thinning and cluster analysis for environmental thinning. Models were compared using AUC, overlap species probabilities in environmental (Denv) and geographic (Dgeo) space. Biased models invariably had lower AUCs than unbiased models and clearly deviated from the unbiased model by all measures. Decrease in AUC, however, was small and the AUC of biased models was usually still in the range generally accepted as a well fit model. The effect size of each bias type depended on the species and evaluation method. The authors focused on Dgeo (Denv was strongly correlated) as the main measure of effectiveness of bias correction. Overall only 29% of all combinations (species*bias type*bias intensity*correction method) showed improvement over the biased model with the simulated species substantially easier to correct (57% were successful corrections). Restricted Background (c) failed in almost all cases (6% successful). All other methods performed better but were differentially ranked depending on the combination of factors. Systematic sampling (a), though not always ranked first, performed most consistently and slightly better overall than the competing methods (33% successful). Bias file (b) and cluster (d) methods sometimes outperformed Systematic Sampling but were slightly less successful overall (23%, 23%). Probably the most important result of this work is the bad performance of the Restricted Background (c) method, as it seems to be consistently used/recommended when fitting MaxEnt to biased data. Though Systemtatic Sampling (a) performs well and consistently, the authors acknowledge that the second main conclusion is that the best way of handling bias is often context specific and so one ought to attempt multiple different correction methods in practice. Their somewhat strangely chosen set of correction methods further reinforces this point as other methods that have been demonstrated as effective went untested or were replaced with minor variants that may have changed their effect.



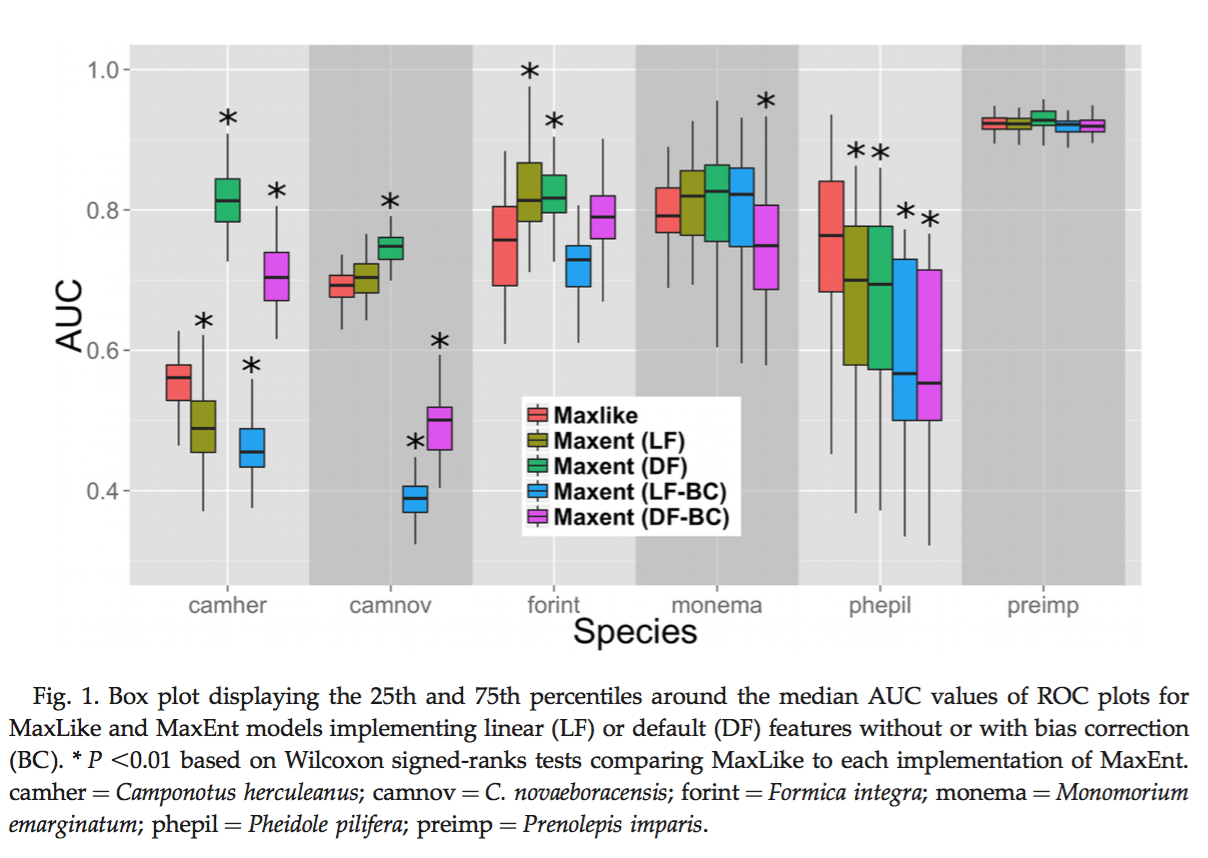 Fitzpatrick, M. C., N. J. Gotelli, and A. M. Ellison. 2013. MaxEnt versus MaxLike: empirical comparisons with ant species distributions. Ecosphere 4(5):55. http://dx.doi.org/10.1890/ES13-00066.1
Fitzpatrick, M. C., N. J. Gotelli, and A. M. Ellison. 2013. MaxEnt versus MaxLike: empirical comparisons with ant species distributions. Ecosphere 4(5):55. http://dx.doi.org/10.1890/ES13-00066.1