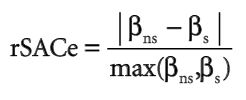Searcy, C. A., & Shaffer, H. B. (2016). Do Ecological Niche Models Accurately Identify Climatic Determinants of Species Ranges? The American Naturalist, 187(4), 1–13. http://doi.org/10.5061/dryad.667g2
A major question surrounding ecological niche modeling is if models accurately reflect the biological ranges of species and if they are informative regarding a species niche requirements. If they do reflect these ecological “truths”, and not simply correlations with climatic variables, then their use in the prediction of species ranges into future climatic conditions is valid. To explore this issue, Searcy & Shaffer (2016) compare climatic variables that determine recruitment in the field with those predicted as high ranking by MaxEnt. Using two decades of demographic data on the endangered California Tiger Salamander, the authors replicated BioClim variables using climate data from nearby weather stations and ran ANCOVA models to measure how well a climatic variable correlated with juvenile recruitment of the salamander. They then created two MaxEnt models:
– a basic model that used permutation importance to rank variable importance
– an informed model that used permutation importance and percent contribution to rank variable importance, and
– corrected for sampling bias
– limited background points based on natural history of the species
– used model selection to select the model’s regularization multiplier
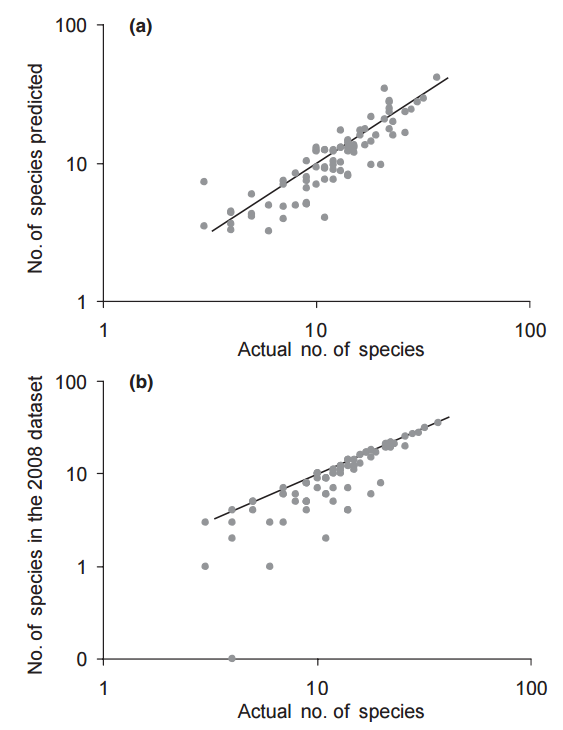
They then compared the variable importance rankings from the ANCOVA models to the two MaxEnt models, evaluating ranking and the response curves, the latter to see if the relationship between the variable and habitat suitability/recruitment was the same.
They found six variables to be highly correlated with recruitment, and these six variables were highly correlated with those predicted as important by MaxEnt, when using the informed model with importance by permutation. Notably, this was not seem with the other models, suggesting that an informed model using permutation may best illustrate biological realism. Interestingly, the response curves were not all the same, with temperature variables exhibiting similar response curves, but precipitation variables correlated in opposite directions. This may be due to the fact that they only considered linear responses, effectively dropping most MaxEnt curves, which were non-linear. It may also be due to the temporal scale of rainfall, in which one year with above-average rainfall can lead to high population growth, but an overall increase in rainfall over many years can decrease population growth.
In general, this paper provides evidence that ENM is based on biological realism, albeit with several caveats. Only the informed MaxEnt model with permutation reflecting this conclusion, suggesting that variable ranking by permutation should always be chosen, and models should be corrected for sampling bias and natural history, and controlled with a regularization multiplier. It also stresses that many biological responses are non-linear, so any models that treat them as such are likely to fail.
Note: They also use the MaxEnt model to predict the effect of climate change on the salamander, but this seemed less relevant and generalizable to the class, so I didn’t report on that aspect.