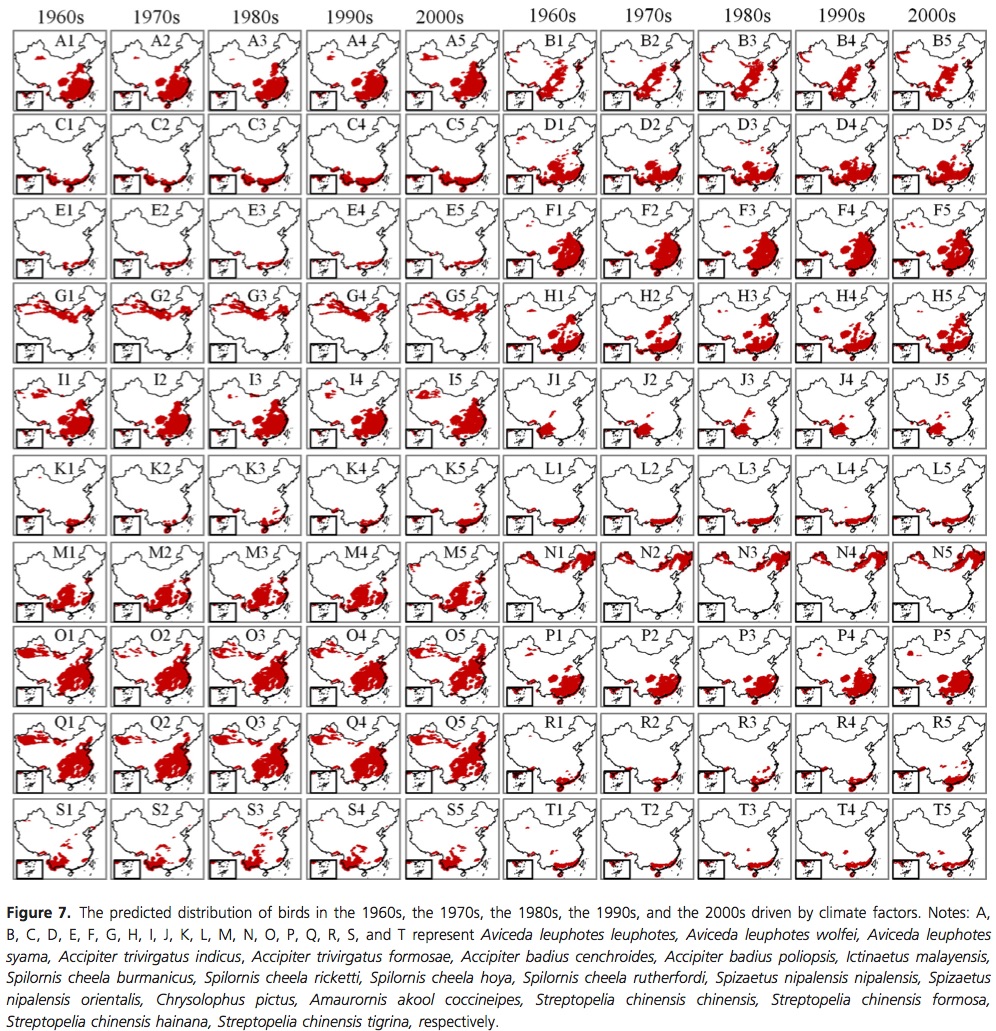
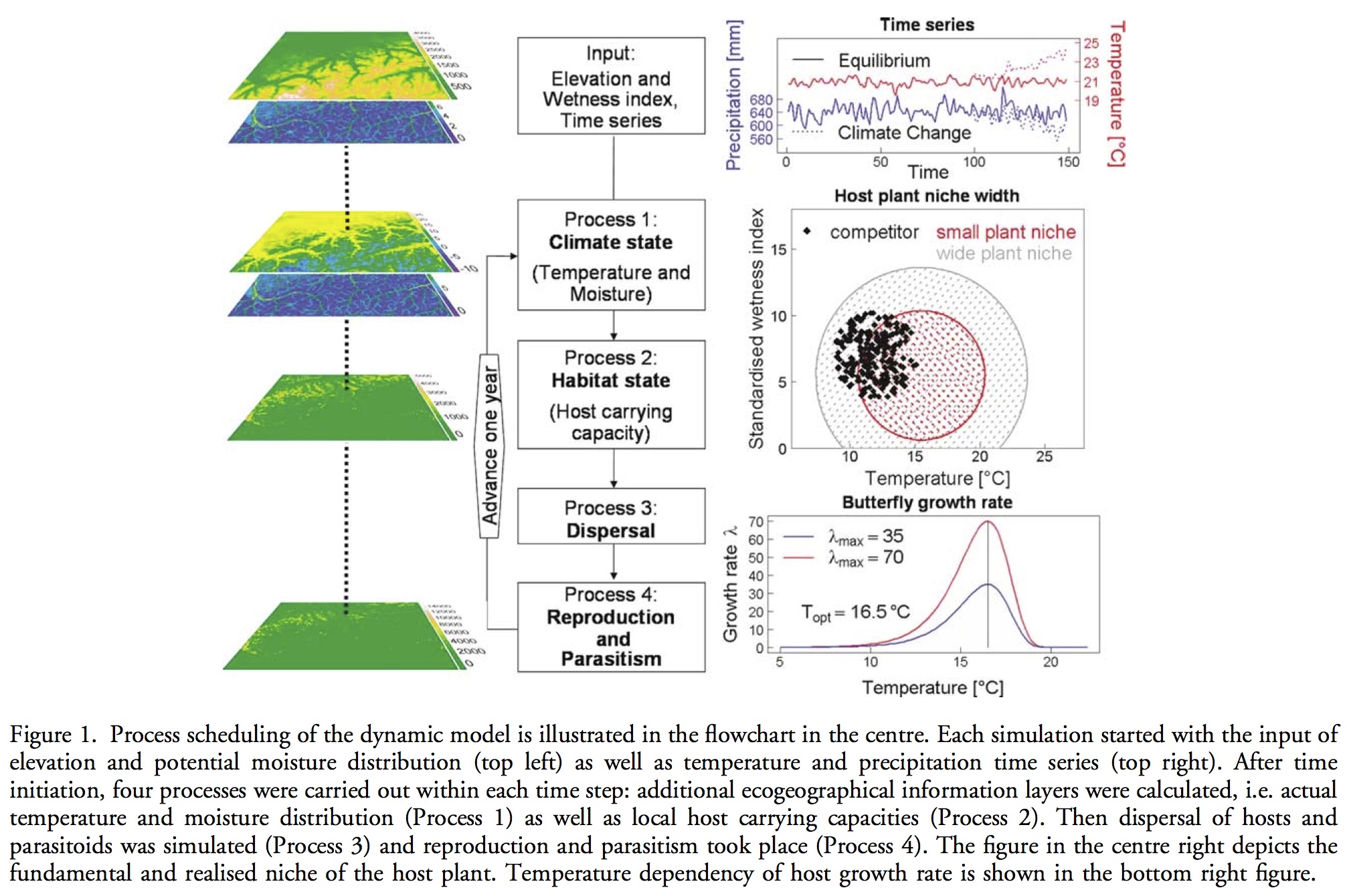
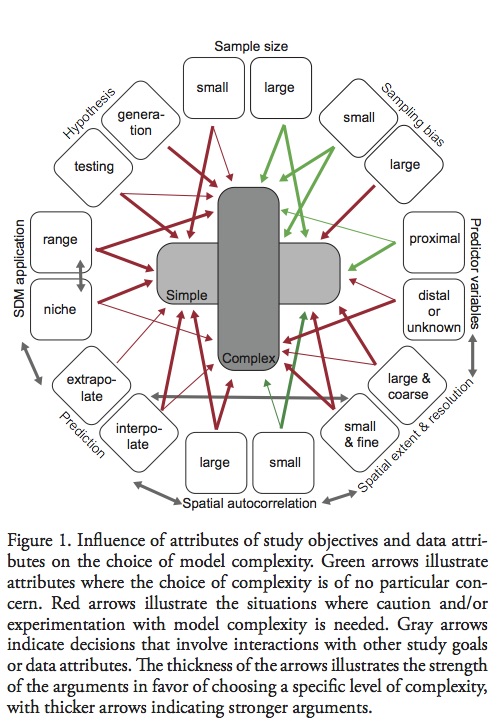
Hawkins, B. A. (2012), Eight (and a half) deadly sins of spatial analysis. Journal of Biogeography, 39: 1–9. doi: 10.1111/j.1365-2699.2011.02637.x
Spatial autocorrelation is not the only issue of spatial analysis. Additionally, this autocorrelation is not just a data quality issue. Issues raised are focused on regression models.
1. Spatial autocorrelation generates bias
Nature is autocorrelated, species are distributed non-randomly. Understanding the pattern in autocorrelation the goal of ecology and biogeographers. However, statistical parametric modeling often requires random data- so perhaps this approach, specifically significance testing, is not appropriate.
2. Spatial regression is best
A common assertion in the literature: If ordinary least square regression is biased, then generalized least square must be the best (and only) method.
But there are multiple ways to to cope with the bias (or uncertainty), there is no single best approach. Alternatives include presenting multiple models or model averaging, however, this will never correct for uncritical use of multiple regression.
3. The world is stationary
Stationarity is the assumption that predictor/response variables are invariant throughout data. The consequences of this violation varies with model choice- but will influence the interpretation of parameter values. Despite non-stationarity being common in ecological data, very few studies test or account for this assumption. This needs to be done at the very least, if ideally the authors do not incorporate non-parametric methods such as CART.
4. Partial regression coefficients mean something
Ecologists would like to identify the most important influence on spatial patterns, but multiple regression is designed to ignore correlations among predictors making this a very poor approach. Alternatives, such as, CART or SEM are better suited to assert causal links.
5. Regression coefficients identify effects
`Correlation is not causation’ is well known, and ignored. The distinction between statistical effect and mechanistic effect need to be clearer in both communication and thought.
6. Species richness generates bias
This is a misunderstanding of sampling theory. All samples will converge to the parametric mean, if the sample is random. The non-random assortment of species are the patterns we are trying to test. The need to correct for species richness is the result of confusion between bias and precision. It is clear that the claim that richness generates bias in estimates of means is without foundation.
7. The earth is round (P<0.05)
P-values and AIC/BIC are not complementary tests for model evaluation. Either the model should be compared to the null (as in p-value) or the most parsimonious model should be chosen (AIC/BIC). CART can lend itself to model selection based on information theory.
8. Spatial processes explain spatial patterns
Legendre (1993) provided a heuristic method for distinguish- ing environmental and spatial structure in ecological data by means of a partial regression (or constrained ordinations) that partitions ‘(a) nonspatial environmental variation’, ‘(b) spa- tially structured environmental variation’ and ‘(c) spatial variation of the target variable(s) that is not shared by the environmental variables’ (p. 1666). His use of the language was careful, and this method is now widely used, but it is not uncommon to read that (c) is the effect of pure space, or the effect of spatial processes. Is it?
8 and half. Spatial autocorrelation causes red shifts in regression models
Overemphasize on the importance of broad scale (vs local) predictors is called a red shift. If anything, we have this backwards. Range maps contain false positives, and survey data contain false negatives. Range maps are created by filling in ‘presences’ between points, meaning that closer cells will have more distortion than distant cells. Of course, the level of distortion is grain dependent, but so are the processes that influence diversity.