Mod, H. K., et al. (2015). “Biotic interactions boost spatial models of species richness.” Ecography 38(9): 913-921.
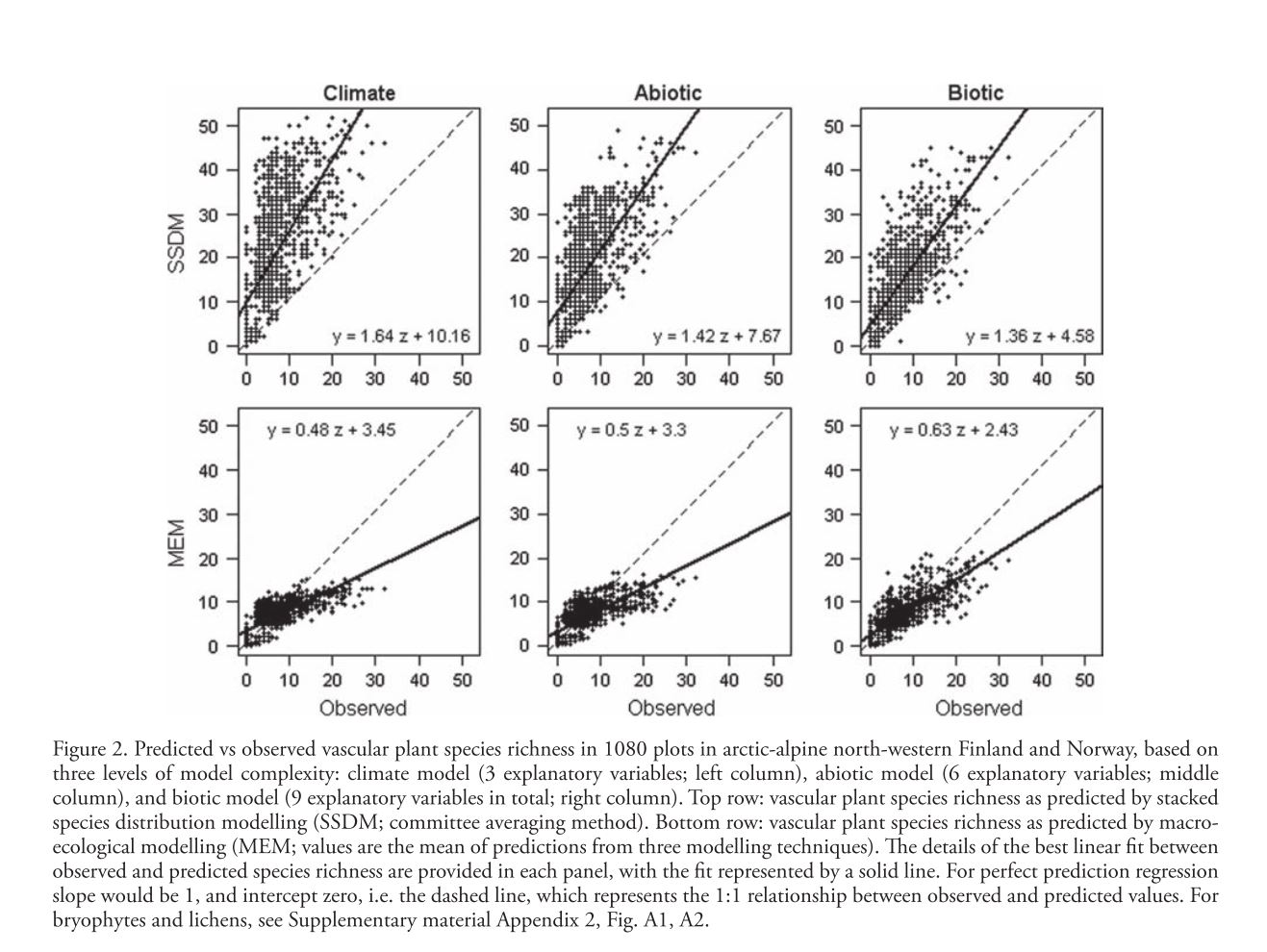
Mod et al. attempt to address the general lack of quantitative consideration of biotic interactions in spatial modeling. Rather than basic spatial distribution modeling they model species richness across a landscape with two different methods. The first, somewhat familiar, method is stacked species distribution (SSDM) in which species distribution models are fitted for all species and then overlaid to determine species richness at each point. These models are fit using Generalized Linear Models (GLMs), Generalized Additive Models (GAMs), and Generalized Boosted Models (GBMs) and SSDMs are generally expected to overpredict species richness because the simple stacking implies no intrinsic environmental carrying capacity. They also use macroecological models (MEM) which directly model species richness and implicitly consider the environment to be limiting to the number of species. MEMs do not make any distinction between different species and generally tend to overpredict richness in species-poor sites while underpredicting richness in species rich-sites. In order to ascertain the ability of biotic variables to improve prediction and potentially correct these problems the authors build 3 different types of models and fit them to 3 taxonomic groups (vascular plants, bryophytes, and lichens). The first (Climate) model includes mean air temperature of the coldest quarter, growing degree days, and ratio of precipitation to evaporation, all broad-scale environmental drivers known to have a strong impact on vegetation. The second (Abiotic) model includes the Climate model as well as soil quality, soil wetness, and solar radiation as finer scale abiotic predictors. Finally the third (Biotic) model includes all previous predictors and the cover of three dominant species known to have impacts on the distribution of other species. These three species show both competition and facilitation based effects on a number of different species. In order to determine the fit of different models a linear regression was fitted to the plot of predicted vs. observed species richness (slope = 1 and intercept=0 represents perfect prediction). The inclusion of biotic variables increased fit and decreased bias for both methods across all taxa, the regression slope and intercept more closely approaching the ideal values. Mean AUC values averaged across all species models built for SSDM were higher as well. The fact that inclusion of biotic variables significantly improved fit across two different modeling methods strongly supports the extra explanatory/predictive power this data can offer. The widespread application of these methods relies, however, on the accurate determination of important biotic variables. This study was able to approximate competition pressure using the cover of 3 dominant species, an assumption which may be generalizable to a number of shade/nutrient limited plant systems. Systems with a more diverse and “evenly distributed” competition landscape may be very difficult to model in this way because knowledge of many species’ distributions across the landscape may be necessary to build these models.