Sehgal, R. N. M., et al. “Spatially explicit predictions of blood parasites in a widely distributed African rainforest bird.” Proceedings of the Royal Society of London B: Biological Sciences 278.1708 (2011): 1025-1033.
—
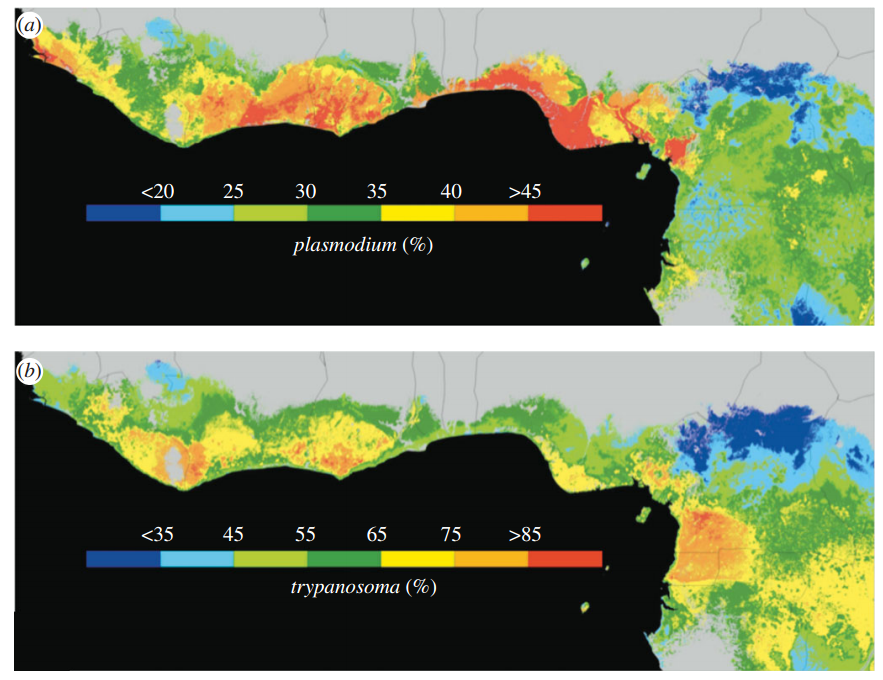
Predicting the potential spatial distribution of parasite species has both obvious rewards (e.g., mitigating human disease) and inherent difficulties. One of these difficulties is that the distribution of parasites is commonly determined by two different, but interacting, filters. Parasite species are obligate at some stage, meaning their distributions are constrained by host distributions. Further, they are still subject to the external environment. Here, the authors use infected host records as point occurrences to train Maximum Entropy models. Specifically, occurrence records consisted of olive sunbird hosts infected by one of two avian parasites (_plasmodium_ or _trypanosoma_). These parasites exist on other hosts, and the host likely exists outside of the area examined (West Africa). Using these occurrence records, they created geographic maps of occurrence probability of infected birds (as that is what their occurrence records are). They determined environmental variable importance (Figure 1 in the paper) for both parasites, and then combined results from a random forest analysis to predict pathogen prevalence across space. This was done by training random forests on prevalence data using environmental covariates, and then projecting the reuslts onto unsampled regions in space, constrained by the MaxEnt occurrence probability predictions. Neat idea, neat paper, lots of questions raised about their approach. There are many assumptions built-in using infected hosts as occurrence points, and even more in projecting prevalence-environment relationships onto point predictions from MaxEnt (i.e., doesn’t this assume transmission is not a function of host density, population genetics, interacting community of hosts/non-hosts, etc., but is instead only a function of environment?).