Motivation:The Pilosocereus aurisetus complex is comprised of 8 cactus species associated with the rocky savannas in eastern Brazil. Species have been defined by morphological and genetic traits. However, different genetic markers lead to different conclusions. For these reasons the authors attempt to answer the following questions regarding the complex diversification:
(1) Are the northern P. aurisetus populations more related to the other conspecific populations in the Espinhaço Mountain range or to population from other species in Central Brazil, as shown by cpDNA data?
(2) Is the currently recognized P. machrisii species composed of two distinct lineages?
(3) What is the relationship of P. jauruensis with the other species of the complex?
Additionally, the authors also tested climatic niche differences between the observed geographic lineages with the hopes of making some inference of the complex’s phylogeographical history.
Methods: Amplicons from AFLP of 40 Pilosocereus samples consisting of 4 species from P. aurisetus species group and and out group. These species have the widest distribution and were the most phylogenetically unresolved. Sequences were processed to identify loci and then alleles across the species and populations. The alleles were used to infer the most likely number of interbreeding groups in the data set without any sampling site information. The most likely number of interbreeding groups were then treated as operational taxonomic units (OTUs) and used to estimate a species phylogenic tree. Species occurrence data was obtained by GPS measurements during transacts of the range in addition to occurrences in the global biodiversity information facility. Sample sizes were generally small for each species and therefore not prone to over fitting. Climatic divergence in addition to genetic divergence was tested by grouping the occurrences according to the genetic lineages recovered by phylogenic analysis. The effects of past climatic oscillations on the niche of each lineage were determined by fitting the models in the present, 21 kya (LGM), and 135 kya (LIG) scenarios using 3 different algorithms. Of the 19 BIOCLIM variables, the authors used 6 which were which were showed to have low correlation and high informativeness. The model outputs were converted into presence/absence data based on a threshold value where the ratio of true positives to actual positives and true negatives to actual negatives is equal. In a area with at least 3 overlapping projections was considered suitable – climactic stable areas were suitable in all 3 time periods.
Results and Discussion: The genetic analysis inferred 5 mating groups split between two main geographic lineages. The two lineages had minimal overlap in all time periods, this overlap was even smaller far stable areas (overlap in all three times). The climatic niche does not appear to have changed over time indicating that range shifts were not crucial for present day distributions.
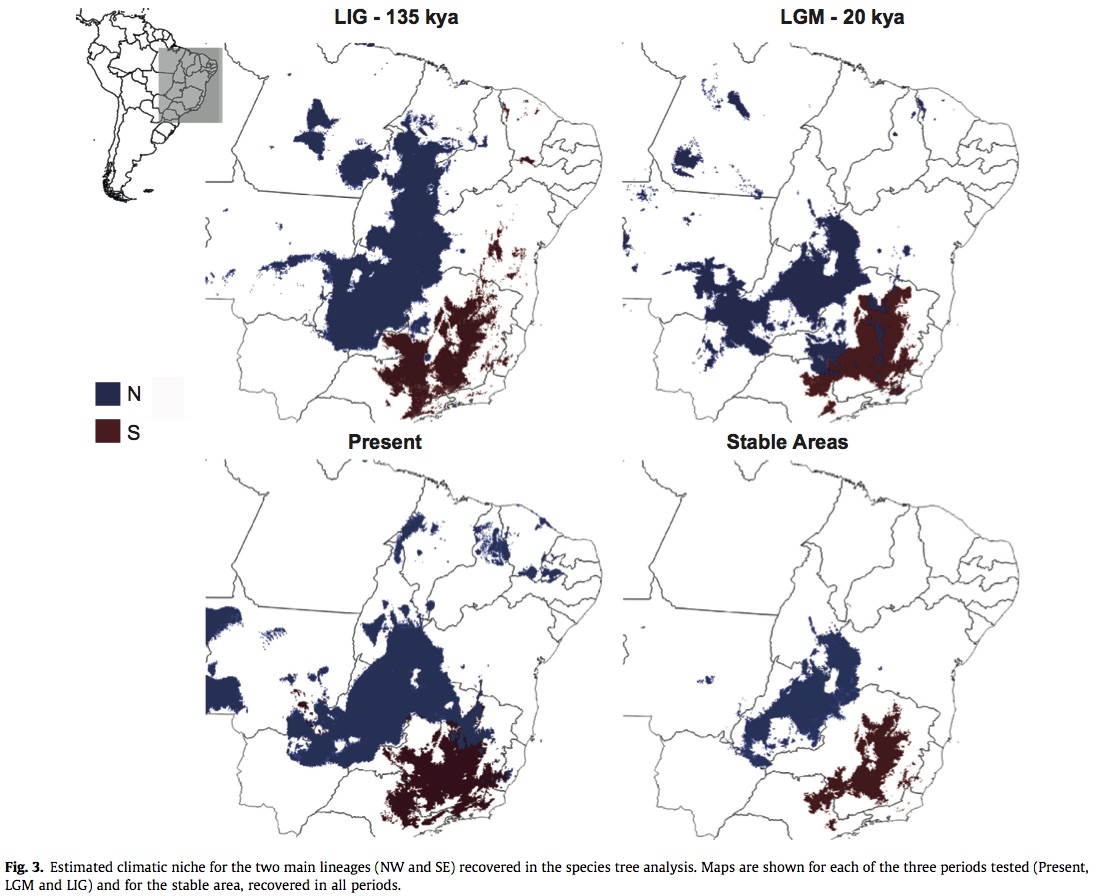
Thoughts: The genetic analysis was very thorough and well developed. However, the niche mapping wasn’t fully integrated into the rest of the study. I think this is a good example of the consequences of developing easy to use data (WorldClim). It is not really clear how the determining the climatic niche over time strengthen the authors’ phylogenetic conclusions.
Manolo F. Perez, Bryan C. Carstens, Gustavo L. Rodrigues, Evandro M. Moraes. Anonymous nuclear markers reveal taxonomic incongruence and long-term disjunction in a cactus species complex with continental-island distribution in South America. Molecular Phylogenetics and Evolution. Volume 95, February 2016, Pages 11–19 doi:10.1016/j.ympev.2015.11.005
