Sandve, Geir Kjetil, et al. “Ten simple rules for reproducible computational research.” PLoS Comput Biol 9.10 (2013): e1003285.
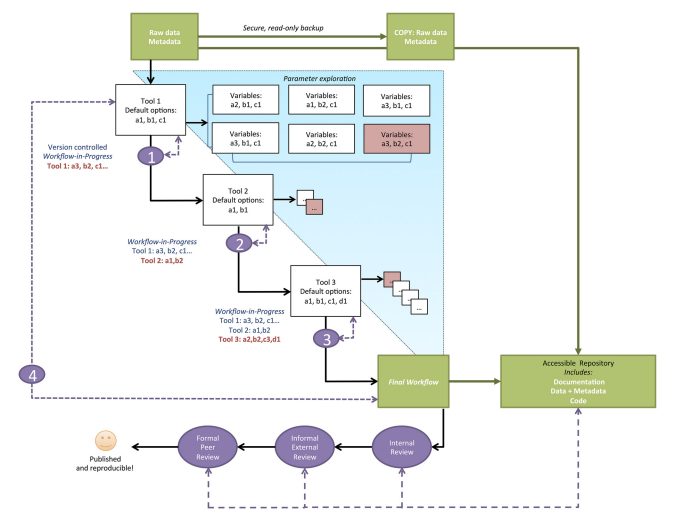
Scientific research has seen a rise in recent years of the increasing demands for more computational skills required to conduct biological and ecological research. This increase in research complexity has precipitated the need to stress the importance of reproducibility of computational methods. Reproducibility is an abstract concept that provides a way to consider how likely someone else provided your data and methods could recreate the results reported in your manuscript. Provided in this paper are ten simple rules that every researcher should consider when conducting a computational based experiment.
First, provide information regarding how every provided result was produced, even if the result ends up not making the final draft. This will not only help recreate the results reported, but also provide insight into the parameter selection process. Secondly, avoid manual data manipulation steps when possible. Data manipulation is a feature of almost any study; however, manually editing data is something that can not be easily recreated with a script. Third, make backups of software versions used in analysis. Packages and programs are updated constantly, and sometimes updated one package may break its dependencies on another. Currently, researchers can use docker as a means to save current software versions. Fourth, version control all custom scripts. This is related to the fourth rule, by version controlling your scripts you are increasing the likelihood that the script will be able to run in the near future. Fifth, keep track of intermediate results. This will help should you ever need to return to an intermediate step in your analysis and make corrections. Sixth, make notations for how to recreate stochastic data. If you induce random noise into the data it’s best to provide the parameter values governing how the noise is manifested. Seventh, store the data behind plots. This cuts out the need to rerun potentially time consuming analysis. Eighth, in your final script save data outputs at identifiable milestone markers within your analysis. Ninth, Where possible provide information regarding why you selected for certain parameters or methods instead of others. Lastly, upload your scripts and data into public domain for ease of access to other researchers.