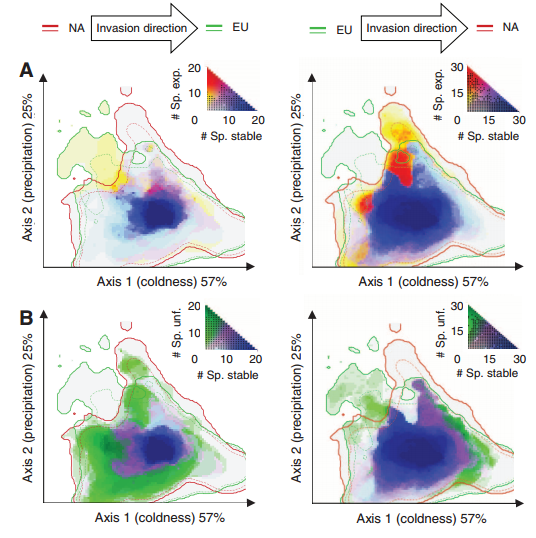
Species may be able to shift their climatic niche requirements according to some researchers, while most models of diversity as a function of global change assume a constant climatic niche for a given species. This is especially the case in species invasions, where the species may be able to exist in climates different from the native range. The authors examined 50 invader plant species in North America and Eurasia in order to quantify the degree of niche expansion, niche overlap, and niche unfilling (species only partially fills its native niche in the invaded range). They found niche conservatism in 46% of the plant species, suggesting that plants largely occupy similar niches in their invaded range. Further, the authors found that when niche expansions were present, 14% of these species show greater than a 10% niche expansion, which they argue is rather low. Niche unfilling was rather common (48% of species), though they don’t discuss the influence of dispersal limitation or the influence of time since invasion on this result. One would think that recent invaders that haven’t had time to disperse (or those under active control) would not have time to reach the extent of their niche, which would appear as niche unfilling compared to their niche in the native range. In their supplement, the authors detail how they calculated the E-space used to quantify niche overlap (a standard PCA whose first two axes explained 82% of the variation in 8 variables including degree day, evapotranspiration coefficients, temperature and precipitation).
Tag: Tad Dallas
Facilitation and the niche: implications for coexistence, range shifts and ecosystem functioning
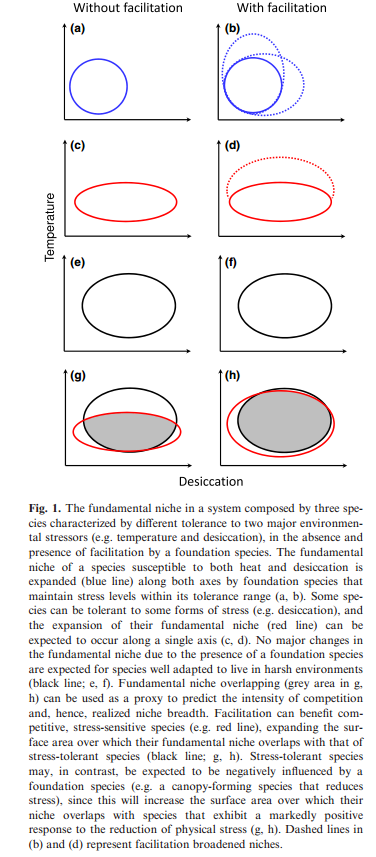
The original description of the niche purposefully excluded the impact of species interactions for niche determination. In a more applied context, several authors have included species interactions, specifically negative interactions, in determining a species niche or geographic range. Here, the authors consider the effect of facilitatory interactions on the expansion of the niche. An example would be if the presence of another species created habitat necessary for the colonization of a secondary species, such as the case with woodpeckers facilitating the presence of tree hole dwelling insects. Species interactions could create suitable environments (e.g., microhabitat variation; see Chesson (2000)) or provide access to a resource that could enable persistence. The authors argue that the introduction of a facilitatory species could result in a niche shift of a focal species in E-space (as in Figure 1). The authors also discuss the role of coexistence when competing species are both responding to a facilitatory species. They argue that niche broadening as a function of facilitation may promote exclusion of one of the species, as niche broadening may put two species in competition where they previously weren’t. This will only be a big issue when fitness differences are large.
Chesson, P. (2000a) Mechanisms of maintenance of species diversity. Annual Review of Ecology and Systematics, 31, 343–366
SDMdata: A Web-Based Software Tool for Collecting Species Occurrence Records
Obtaining data dynamically and programmatically is necessary for reproducible research. This a blanket statement. What I mean specifically is that the ability to access data programmatically from a source that is version controlled allows for the consistent use of data. Currently, many databases are accessible through web-based interfaces, but have no API or method to access the data programmatically. This matters because subsequent analysis of the data is based only on that snapshot from a potentially dynamic database. Ideally, a complete workflow would include pulling the data from a database, cleaning it, analyzing it, and outputting results. This paper introduces a tool to download and clean species occurrence data from GBIF (Global Biodiversity Information Facility). This tool is web-based, written in Python, that takes a species name list, and outputs occurrence data from GBIF. They argue that the current _R_ implementation (`rgbif`) is flawed because of memory limitations (which is a pretty facile argument). I do like that `SDMdata` has an error-checking feature that will flag suspected errors. However, the proliferation of tools to query databases tends to “muddy the waters” in my opinion. Several resources already exist for programmatic data acquisition from GBIF in R, SQL, and Python. Perhaps this tool adds something novel; perhaps we should focus on making existing tools better.
Not the time or the place: the missing spatio‐temporal link in publicly available genetic data
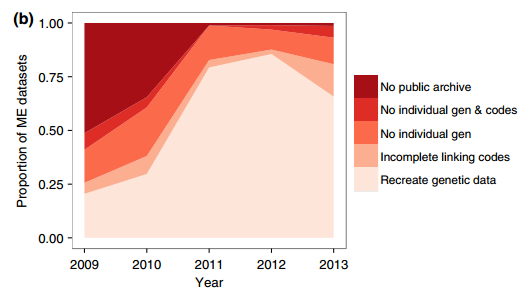
Data archiving is mandatory for many journals in order to encourage data openness, and the re-use of scientific data. However, **how** the data are archived can be really important in determining the usefulness of the data to researchers. Further, data archiving itself does not ensure that the study which the data was used is reproducible. This article demonstrates that 31% of genetic datasets archived as a condition for publication in _Molecular Ecology_. This was largely a metadata problem, in that the data and metadata were not linked well. Furthermore, the quality of the data was a bit coarse in some instances, with geographic data provided in terms of geopolitical location instead of geographic coordinates. Taken together, the authors stress that the data deposition policy has promoted the re-use of data, and the quality of data has increased from 2009 – 2013, but that current genetic data formats that do not allow the inclusion of metadata should be revised, and that data should well-documented and curated in an appropriate repository instead of as a supplementary file.