Guo, Qinghua, Maggi Kelly, and Catherine H. Graham. “Support vector machines for predicting distribution of Sudden Oak Death in California.”Ecological Modelling 182.1 (2005): 75-90.
Recently, several types of oak trees in California have been severely impacted by the emergence of Sudden Oak Death, an infectious disease caused by the pathogen Phytophthora ramorum. Using support vector machine (SVM) approach, researchers provide a prediction for the distribution of sudden oak death with both two class and one class svms.
Researchers argue that a presence only modeling approach, with SVM as an example, will increase the prediction accuracy compared to methods that use a pseudo-absence approach drawn from the underlying distribution of the presence data. Traditionally, SVMs were designed for two class classification for positive and negative or presence and absence for SDM purposes. However, true absence data is often hard to come by. However, a one class, presence only approach, will have a harder time detecting which environmental features are important in predicting the outcome. To overcome this a one class SVM approach was developed.
The training data for this paper consisted of locations where the occurrence of P. ramorum was confirmed in oaks located in California. Host distribution was generated through Landsat ThemP analysis project which provides information at a fine spatial scale (1:100,0000). 14 Environmental variables were used to train the models, environmental information was provided from Daymet. A five-fold cross-validation method was used to evaluate model accuracy.
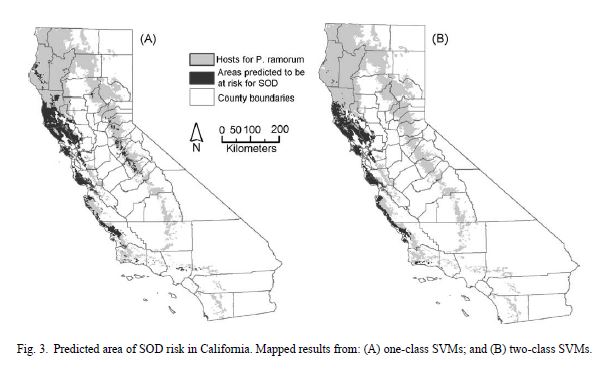
Researchers reported the true-positive rate for your one class SVM was 0.9272 + 0.0460 over an area of 18,441 km2. For the two class SVM reported a true-positive rate of 0.9105 + 0.0712 with a predicted area of 13,828 + 1316 km2. One class SVMs have two main advantages compared to other presence only modeling approached. First, they are able to utilize unique shapes of distributions in feature space through kernel functions. Second, one class SVMs make no assumptions about the distribution of the environmental parameters. Differences in the predicted areas between the two models may indicate that either the one class model has over predicted the area or risk or the two class model has underpredicted. Observed differences can be explained by the higher-true positive rate from the one class model, often false positive rates will increase with the true-positive rate. Another reason for larger risk areas in one class models can be attributed to the two-class model sampling pseudo-absences from presence points, resulting in a more conservative risk estimation. This study demonstrates how a support vector machine approach can be used to ascertain the potential risk of an infectious disease epidemic.
