Shade, Ashley, and Tracy K. Teal. “Computing Workflows for Biologists: A Roadmap.” PLoS Biol 13.11 (2015): e1002303.
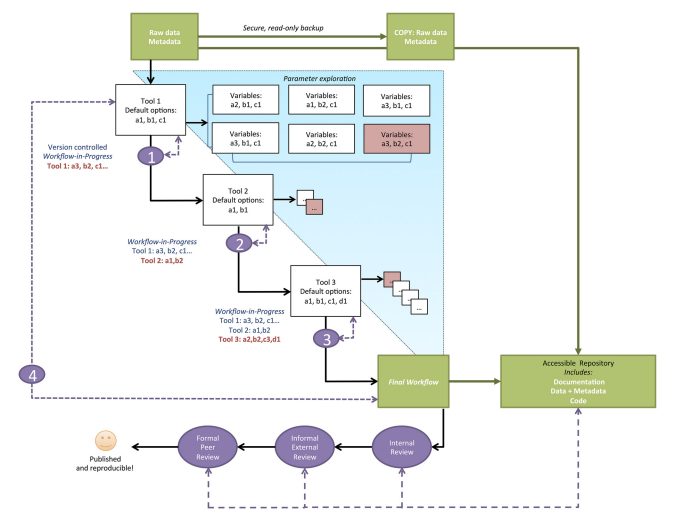
This paper provides a computational framework for biologist in an effort to speed up the development of computational skills needed in contemporary biological research. Broadly the roadmap provided can be broken down into two categories based on within group review and external review. The first step in the computational workflow is to create backup copies of the raw data and metadata and make notes on any data filtration applied before receiving the data. Next, the researcher will want to identify her goals of the study and distinguish whether or not she is conducting a hypothesis test or data exploration. After the goals are properly identified the researcher will want to consider the parameter space which is comprised of all decisions involved in modeling the data (including program selection). Authors encourage adapting branching pattern approach at this point and evaluating parameter space in three key areas: sensitivity analysis, sanity check, and control analysis. Sensitivity analysis observes how model outputs change with change in input options. Sanity checks are inquiring if the model outputs are what the researchers expect to observe or do these results make biological sense. Control analysis uses simulated or provided data to have a firm understanding of the employed model. At certain points in the workflow the researcher will want to conduct reproducibility checkpoints by making sure that given a clean start they can recreate the current step in their analysis. Lastly, researchers will want to utilize online repositories to both backup their data and solicit outside feedback in their approach.