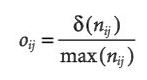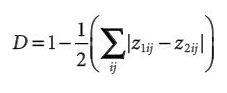Dormann, Carsten F. “Effects of incorporating spatial autocorrelation into the analysis of species distribution data.” Global ecology and biogeography 16.2 (2007): 129-138.
This review paper investigates the importance of incorporating the effects of of spatial autocorrelation (SAC) into any species distribution model. The author was interested in answering two questions. First, does SAC the parameters estimated from species distribution data? Second, does incorporating SAC increase model performance?
The literature review was conducted using Web of Science, search methods the author believed to be reasonable to handle SAC included the following: autologistic regression, generalized least square regression, and correction of significance levels. The search parameters provided to web of science were: “spatial autocorrelation” and “ecology or distribution”, additionally the author would review any papers not returned through Web of Science, but cited in a paper found through the search criterai. The inclusion criteria were: (1) a species distribution was analyzed (2) presence of a traditional analysis (GLM or GAM) and spatial model (3).
Information extracted from the reviewed studies.
|
Arrangement of samples |
Size of neighborhood |
|
Spatial extent/grain |
Type of autoregressive function |
|
Species/group |
Quality of SAC removal/control |
|
Response variable |
Model coefficients |
| Statistical methods |
Importance of SAC |
To measure the effect of correction for SAC the following equation was provided. Where S stands for spatial coefficients and NS stands for non-spatial coefficients.
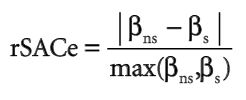
The effect of correcting for SAC on overall model quality was quantified with AIC, R2, and deviance-based pseudo-R2.
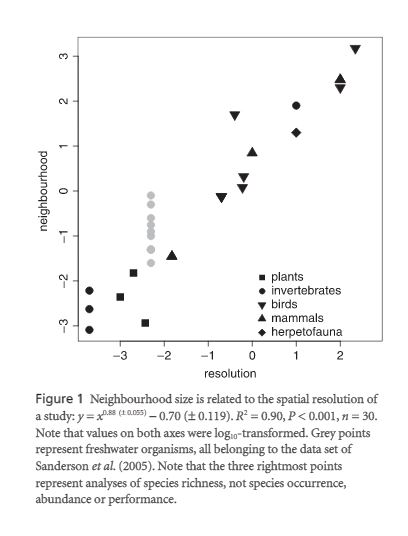
Findings from this study indicate that there was no difference in response type for single species studies in terms of rSACe. The author did find an effect for the range of spatial autocorrelation (neighborhood) and spatial resolution. Meaning that when controlling for the effect of spatial resolution in the study, the effect of SAC was significant. For the effects of spatial autocorrelation on model quality, the author observed a significant improvement in AIC values when SAC information was provided to the model.