Kearney, M. (2006), Habitat, environment and niche: what are we modelling?. Oikos, 115: 186–191. doi: 10.1111/j.2006.0030-1299.14908.x
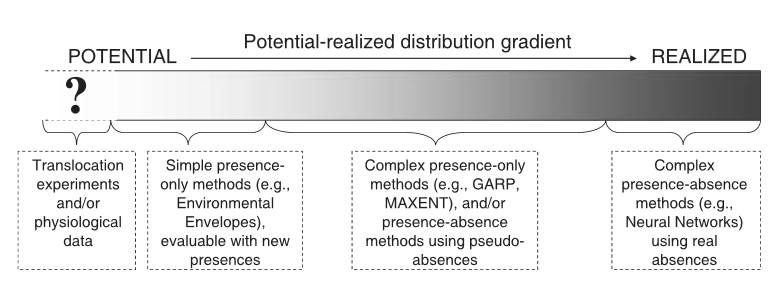
The following words are often used interchangeably when discussing a species distribution: habitat, environment, and niche. These words have been considered “plagued by loose and inconsistent applications of the concepts in describing different methodological approaches” and this paper implies that there is an overall confusion of what is actually being modeled. Generally, the concept of habitat in species distribution is described as being associated with descriptive/correlative analysis of the environments of the organisms and useful in statistical models of species distributions and abundances, while the niche concept is reserved for mechanistic analysis of how different environmental factors in an organism’s habitat interact with the organism itself. While distributions can be potentially predicted by modelling descriptions or correlations between organisms and habitat components, it is suggested to model an organisms niche mechanistically to fully understand and explain distributional limits. This is particularly true when trying to predict an organism’s distribution under altering scenarios, such as climate change. ‘Environment’ would refer to the biotic and abiotic phenomena surrounding and potentially interaction with an organism. Two ways to model species distribution taking into account these “confusingly interchangeable words”, are to 1) use the correlative approach, distribution data and GIS habitat data are associated statistically, often in the form of a regression model then interpolated across regions for which spatial data is available to predict areas of high probability of occurrence (use of the habitat concept) and 2) the mechanistic approach (use of the niche concept) in which the interaction between the properties of the organism and the environmental conditions surrounding it are mechanistically modelled and mapped onto the landscape. Although this paper seems like basic definition issues, these words can often be interchangeably used across different published papers which could be confusing for readers who may be just grasping the ideas and concepts. Thus, this calls for more consistent and universal use of such words to avoid confusion and to pay more attention to the definition and the context.