Marcer, A., Mendez-Vigo, B., Alonso-Blanco, C., & Pico, F. X. (2016). Tackling intraspecific genetic structure in distribution models better reflects species geographical range. Ecology and Evolution, 6(7), 2084-2097.
DOI: 10.1002/ece3.2010
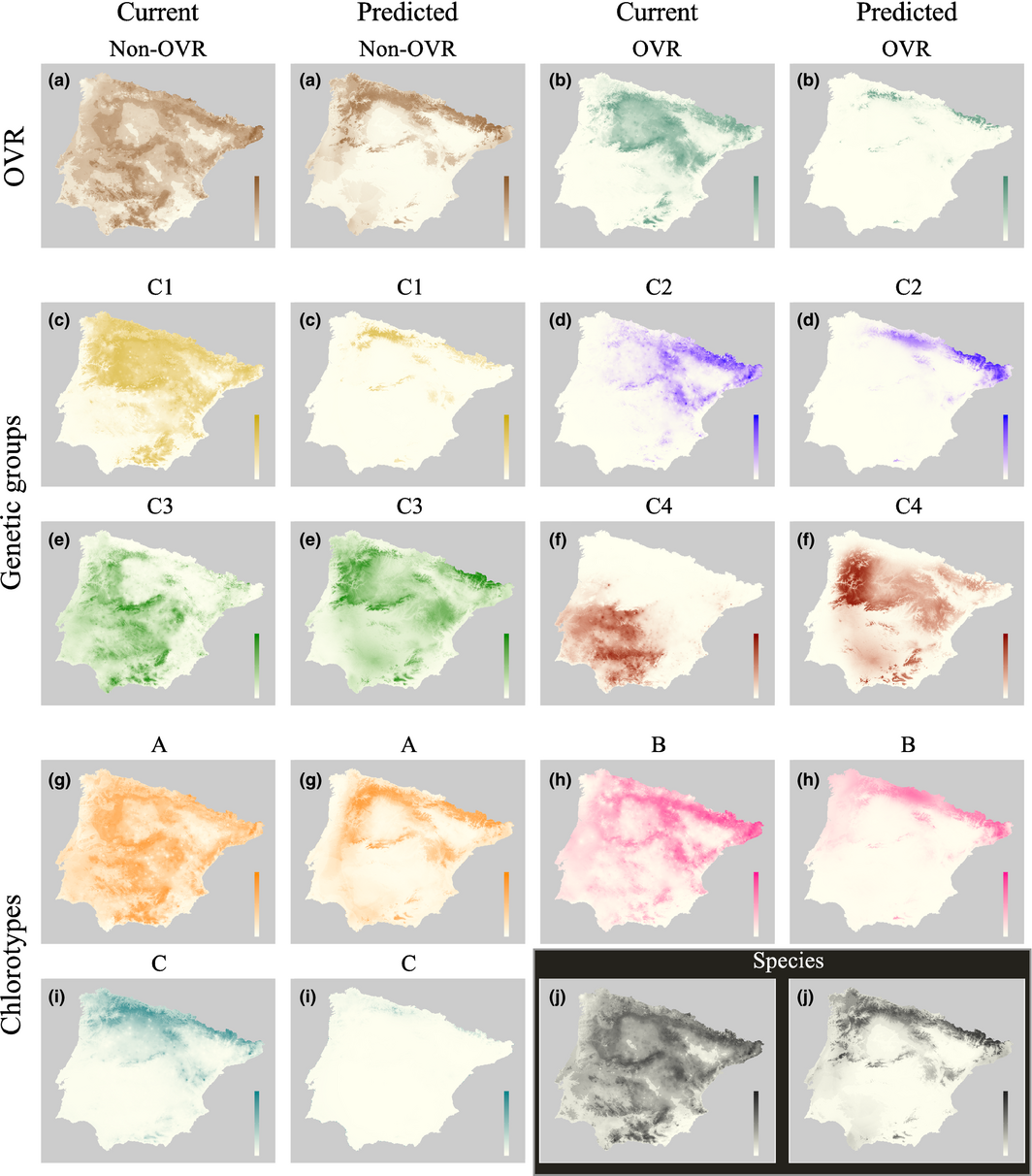
Classical taxonomic designations may not represent the ecological or evolutionary unity that matter to understand the mechanisms that shape biogeographic patterns. Interspecific genetic variation can influence how a species responds to it changes in its environment. This may make genetic variation important in understanding how species distributions will respond to climate change. Work in the Iberian Peninsula has demonstrated that genetic variation in A. thaliana is geographically structured due to the locations of ice age refuges. This study combines SDM with genetic analyses of the plant to determine how SDMs predict the current distribution of A. thaliana genetic units and which environmental variables account for the genetic variation. The species was split into four genetic clusters on the Iberian Peninsula. A MaxEnt model was constructed using three environmental data sources and 279 presence records which correspond to the genetic samples. The distribution of the species was mainly explained by pH and agricultural land, with the species more likely to occur in acidic conditions. When the model is broken down by cluster variable importance changes from model to model. This highlights the importance of incorporating genetic variation into the predictive model. These units vary in which variables are most important in determining habitat suitability and as the environment changes the species may be influenced in different way throughout its range. By separating the species into distinct genetic units we may be able to understand fluctuations in species distribution better as climates change.

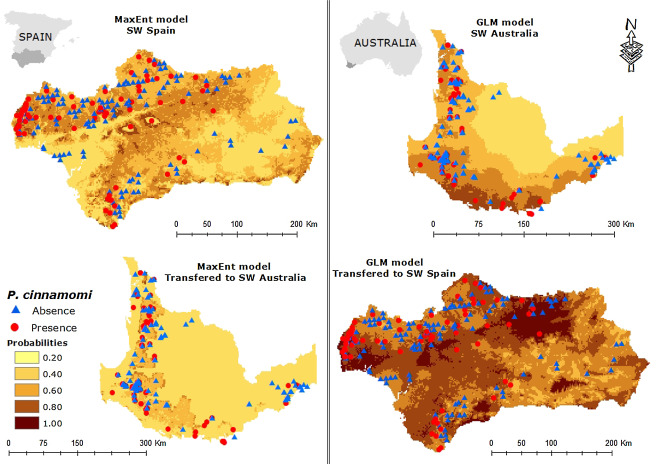 Duque-Lazo, J., van Gils, H., Groen, T. A., & Navarro-Cerrillo, R. M. (2016). Transferability of species distribution models: The case of Phytophthora cinnamomi in Southwest Spain and Southwest Australia. Ecological Modelling, 320, 62-70.
Duque-Lazo, J., van Gils, H., Groen, T. A., & Navarro-Cerrillo, R. M. (2016). Transferability of species distribution models: The case of Phytophthora cinnamomi in Southwest Spain and Southwest Australia. Ecological Modelling, 320, 62-70.