Anderson, Robert P., Daniel Lew, and A. Townsend Peterson. “Evaluating predictive models of species’ distributions: criteria for selecting optimal models.” Ecological modelling 162.3 (2003): 211-232.
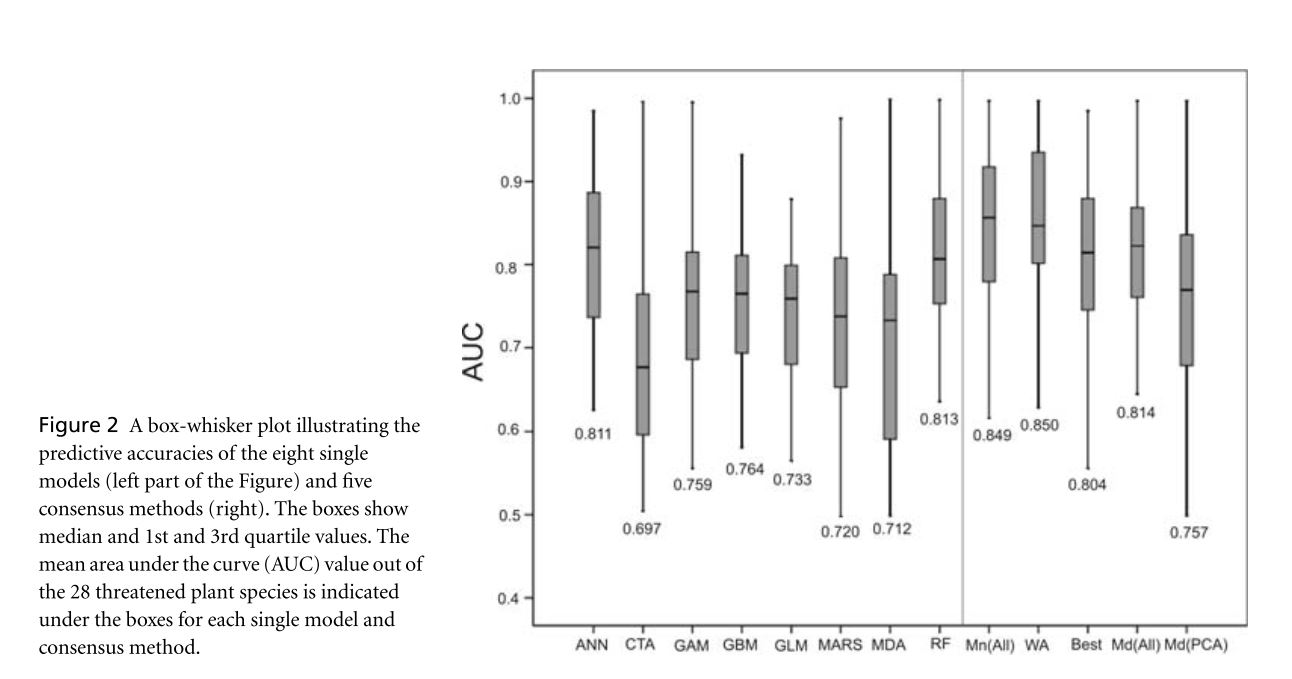
Anderson et al. assess the utility of consensus based predictors in species distribution models. These consensus predictors are made up of a number of fitted species distribution models of varying types. Component SDMs used for consensus modeling were GLM, GAM, MARS, ANN, GBM, RF, CTA, and MDA. These individual models were trained and evaluated on appropriate sub-subsets of the 70% training data subset in order to pre-evaluate these models for the purpose of consensus modeling. Consensus models assessed include Median(All) and Mean(All) which use the median and mean, respectively, of the predictions of all 8 models. The WA approach determines the 4 models with highest accuracy for a given species and computes a weighted average of their outputs. Median(PCA) is calculated as the median of the 4 models for which the variance of the predictions along the 1st principle component of a PCA was the greatest. Finally, Best simply selects the best individual model based on the highest pre-evaluated AUC value. Each of these methods, as well of each of the individual models, were then evaluated using the 30% testing data subset. WA and Mean(All) provided significantly more robust predictions than all single models and all other consensus methods. WA was the best model with a mean AUC of .850 and better predictive performance than all single-models on 21 of 28 species. These methods provide a functional alternative to thorough single-model evaluation and comparison. The fact that the true consensus models consistently outperform the “Best” consensus model suggests the utility of these methods over comparative evaluation. These consensus models also effectively address the common issues that some single-models provide better predictions for interpolation and some for extrapolation and that the best evaluated model often varies significantly from species to species.