Graham, Catherine H., et al. “The influence of spatial errors in species occurrence data used in distribution models.” Journal of Applied Ecology 45.1 (2008): 239-247.
http://onlinelibrary.wiley.com/doi/10.1111/j.1365-2664.2007.01408.x/full
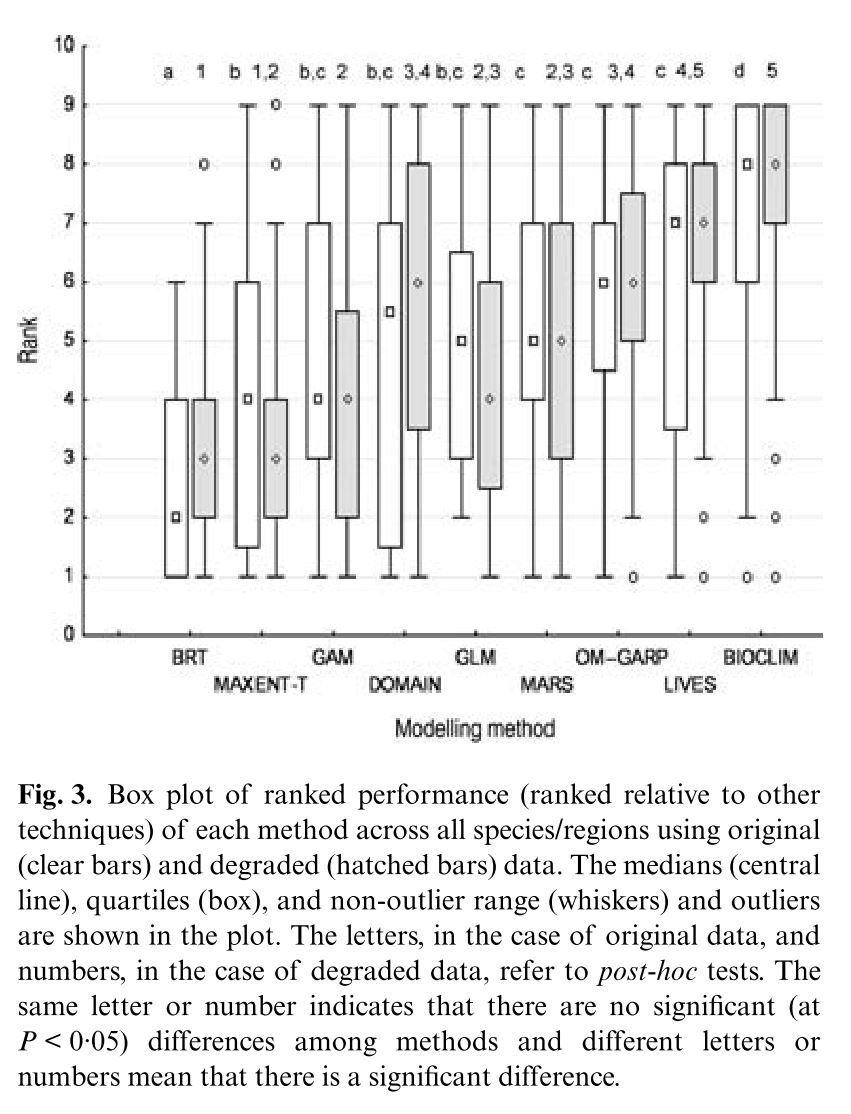
Graham et al. set out to determine what effect spatial error in species presence data can have on a spectrum of Species Distribution Models. Error in species location data can be produced by mistakes in recording or copying of information and broad or imprecise locality information which can be difficult to accurately georeference. Although some of these erroneous points can be identified and removed in data cleaning, this decreases the sample size of training points and in turn the potential accuracy of predictive models. The authors used a data set consisting of 4 geographic regions each with extremely accurate presence/absence data for 10 species. All models were trained using a subset of this data including exclusively presence points (to simulate the typical lack of reliable absence data in museum collections and the like) as well as a version of this data set manipulated such that the x and y coordinate of each presence point was shifted in a random direction by an amount sampled from a Normal distribution with a mean of 0 and a standard deviation of 5km. Area under the receiver operator curve (AUC) was used as a measure of fit of each model, tested against a held-out presence absence data set. Models were directly compared using ranked AUC (i.e. for a specific species and treatment the model with the highest AUC was given rank 1 etc.) in order to account for the fact that direct comparisons of differences in AUC can be a questionable metric. Models tested fell into a few distinct categories, Presence-only models (BIOCLIM, DOMAIN, LIVES), regression based presence-pseudoabsence models (Generalized Additive Models, Generalized Linear Models, Multivariate Adaptive Regression Splines), and relatively new machine learning based approaches Maximum Entropy and Boosted Regression Trees. In general model performance across all region was lower when trained on the error-manipulated data than when trained on the accurate data. There were, however, a number of instances when a model trained on the error-added data performed better than its non-error counterpart. The smallest effect of error on performance occurred in the Australian Wet Tropics where AUC values were relatively low in general and often close to random meaning that not much decrease in performance could be expected. The predictions made by all presence only models, along with GARP and BRT declined significantly with the addition of error. Nonetheless BRT was consistently the best performing model across both data sets (though it was not significantly different from MaxEnt on the error-added data). BIOCLIM and LIVES were consistently the lowest performing models. Presence-only techniques likely suffered the most from added error because they did not have the benefit of the randomly sampled background points with which to weight their models. The authors recognize that this is a useful but relatively limited study with only one spatial data degradation treatment and suggest a number of potential avenues for advancement of this research. Beyond simply increasing the number of different treatments they highlight the need for study of the effects of error in environmental variables used in models and potential methods of mitigating the effects of such error. Although certainly in need of extension and more systematic clarification this study provides some comfort that, even in the face of inaccurate spatial data, many of our preferred modeling methods will only slightly decrease in performance.