A variety of methods can be used to generate Species distribution models (SDMs), such as generalized linear/regression models, tree-based models, maximum entropy, etc. Building models with an appropriate level of complexity is critical for robust inference. An “under-fit” model will introduce risk of misunderstanding factors that shape species distribution, whereas an “over-fit” model brings risks inadvertently ascribing pattern to noise or building opaque models. However, it is usually difficult to compare models from different SDM modeling approaches. Focusing on static, correlative SDMs, Merow et. al. defined the complexity for SDM as the shape of the inferred occurence-environemnt relationships and the number of parameters used to describe them. By making a variety of recommendations or choosing levels of complexity under different circumstances, they developed negeral guidelines for deciding on an appropriate level of complexity.
There are two attributions determining the complexity of inferred occurrence-environment relationships in SDMs: the underlying statistical method (from simple to complex: BIOCLIM, GLM, GAM, and decision trees) and modeling decisions made about input and settings. As for modeling decision, larger numbers of predictors are often used in machine-learning methods instead of traditional GLM. Incorporating model ensembles and predictor interactions will also increase model complexity.
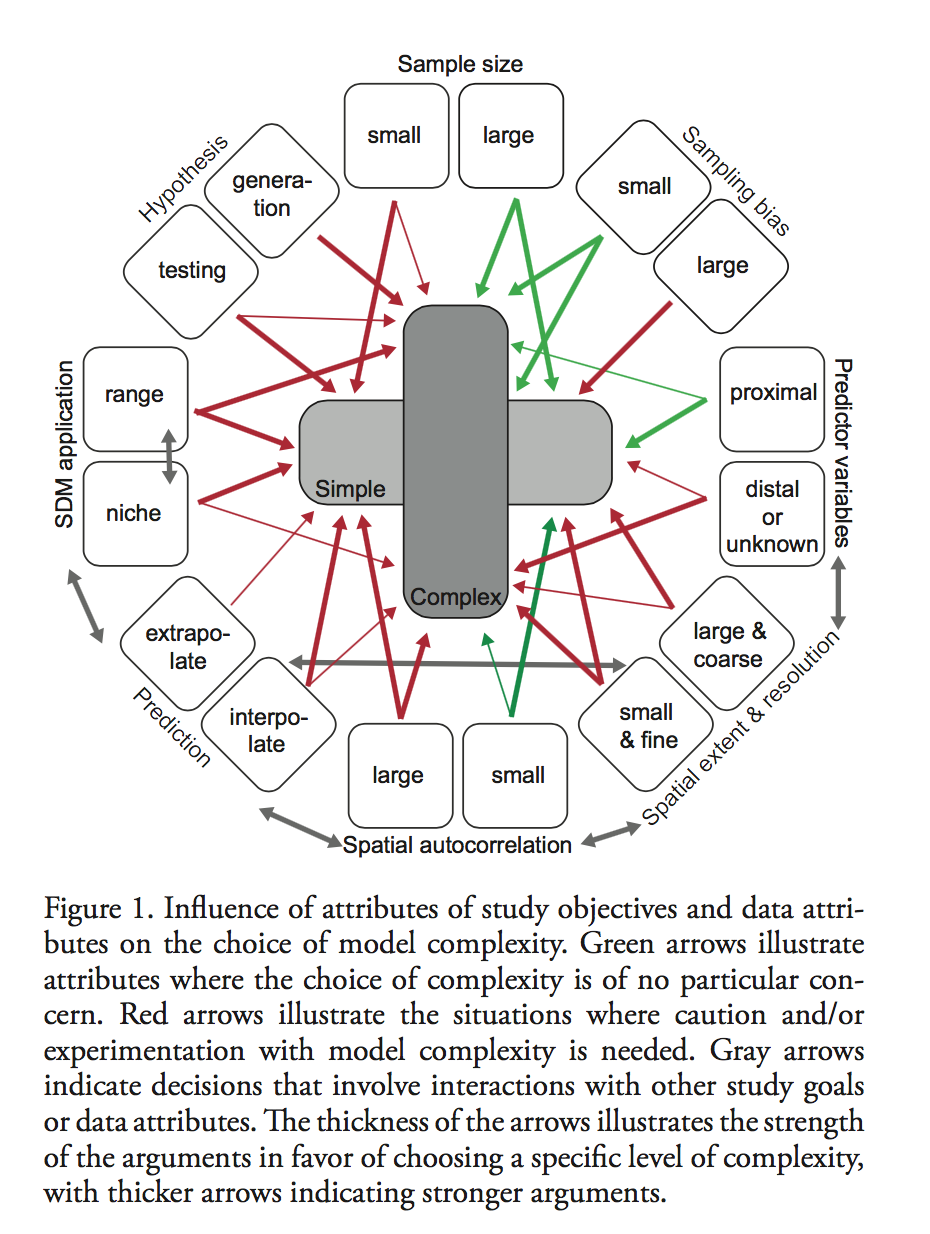
Figure 1 summarized their finding in terms of general considerations and philosophical differences underlying modeling strategies. They suggested that before making any decisions on model approaches, researchers should experience both simple and complex modeling strategies, and carefully measure their study objectives (Niche description or range mapping? Hypothesis testing or generation? Interpolate or extrapolate? ) and data attributes ( sample size, sampling bias, proximal predictors and distal ones, spatial resolution and scale, and spatial autocorrelation). Generally speaking, complex models work better when objective is to predict, and simpler models are valuable when analyses imply only certain variables are needed for sufficient accuracy. Finally, they concluded that combining insights from both simple and complex SDM approaches will advance our knowledge of current and future species ranges.
Relating this paper with the Breiman paper, Merow et. al. regarded data modeling models simpler than algorithmic models, which are usually semi- or fully non-parametric. But they also acknowledged that this conception is rather relative: the interpretability of complex models is not necessarily difficult, and the complexity can still identify simple relationships. However, it can be seen that Merow et. al. regarded interpretation of models one of the goals of modeling. They think there is no absolute situations that simple models or complex models violate the nature of science, but their merits are more case-dependent. I think the combing of simple and complex models, as Merow et al suggested, is the trend of statically modeling, and Breiman maybe over-emphasized the distinctions between the two cultures in modeling world.