Kampichler, C., Wieland, R., Calmé, S., Weissenberger, H., & Arriaga-Weiss, S. (2010). Classification in conservation biology: A comparison of five machine-learning methods. Ecological Informatics, 5(6), 441–450. http://doi.org/10.1016/j.ecoinf.2010.06.003
Machine learning methods have recently been adopted by ecologists to use in classification (eg. bioindicator identification, species distribution models, vegetation mapping) and there is an increasing amount of literature comparing the strengths and weaknesses of different machine learning techniques over a variety of applications. Kampichler et al add to this base of knowledge by comparing five machine learning techniques against the more conventional discriminant function analysis in their application to an analysis of abundance and distribution loss of the ocellated turkey (Meleagris ocellata) in the Yucatan Peninsula. They used data on turkey flock abundance (including absences) from the study area and 44 explanatory variables, including prior turkey abundance in local and regional cells, vegetation and land use types, and socio-demographic variables.
The techniques investigated were
– Classification trees (CT): uses a binary branching tree to describe the relationships between explanatory and predictor variables
– Random forests (RF): constructs many trees and then bags the trees to select the explanatory variables
– Back-propagation neural networks (BPNN): creates a network whose nodes are weighting by the training data
– Automatically induced fuzzy rule-based models (FRBM): processes variables based on algorithms using fuzzy logic
– Support vector machines (SVM): maps training data into an n-dimensional hyperplane and applies a kernel function to maximize seperation between the classes
– Discriminant analysis (DA): combines the explanatory variables linearly in an effort to “maximize the ratio between the separation of class means and within-class variance”
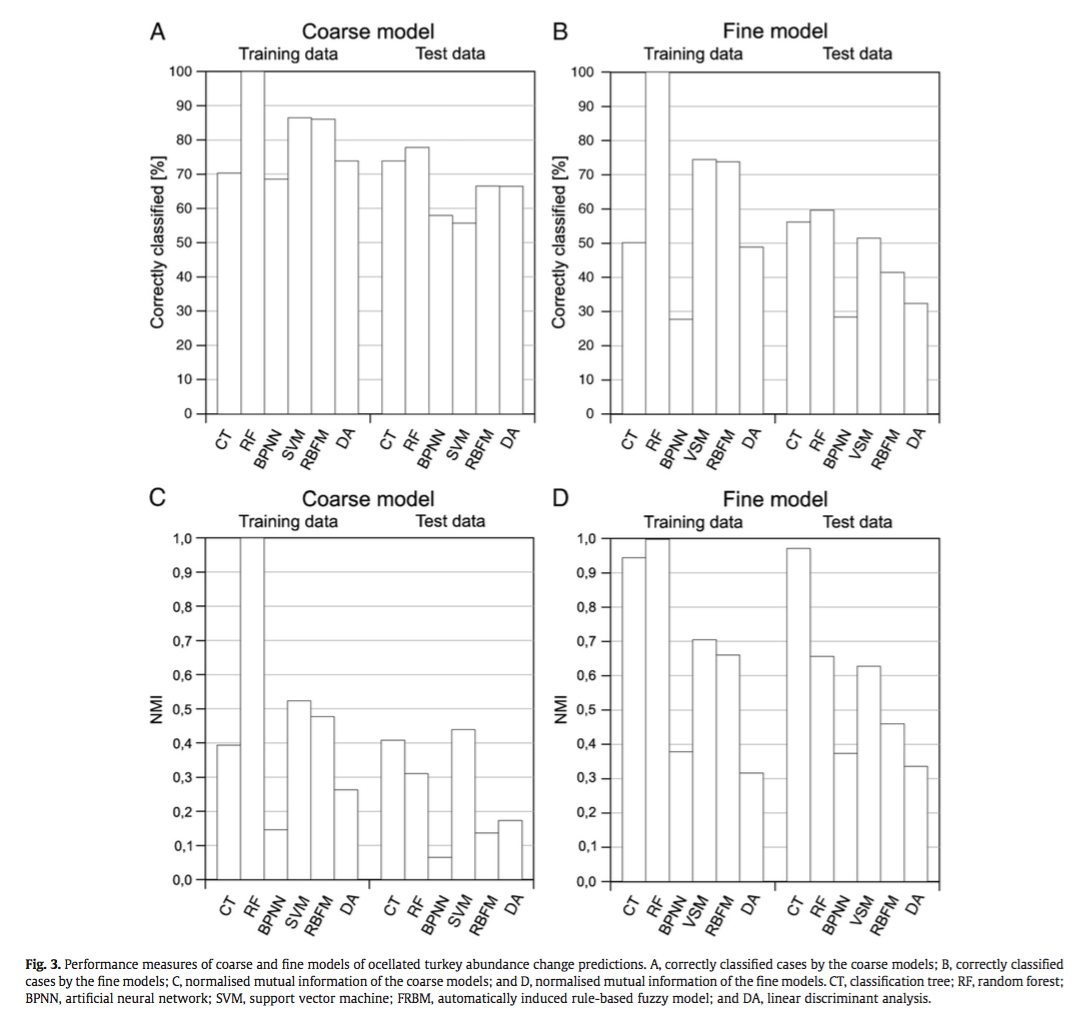
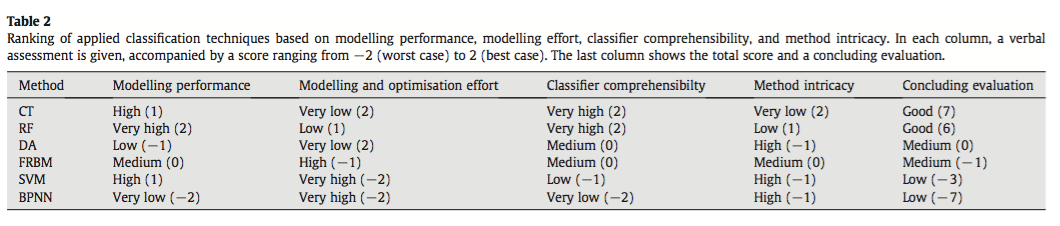
They compared the techniques based on their ability to correctly classify training and test data and using the normalized mutual information criterion, which is based on the confusion matrices and measures similarities between predictions and observations from 0 (random) to 1 (complete correspondence). In general, RF and CT performed the best, however the authors ranked CT first because of its high interpretability. An interesting point brought up is the fact that, in spite of the recent influx of machine learning in the scientific literature, most conservation decisions do not consider their results, most likely because of the lack of their interpretability and expertise needed to optimize the models. With this in mind, SVM, which performs relatively well, may not be the appropriate choice for conservation management because they are not well understood by ecologists lacking the proper mathematical training.