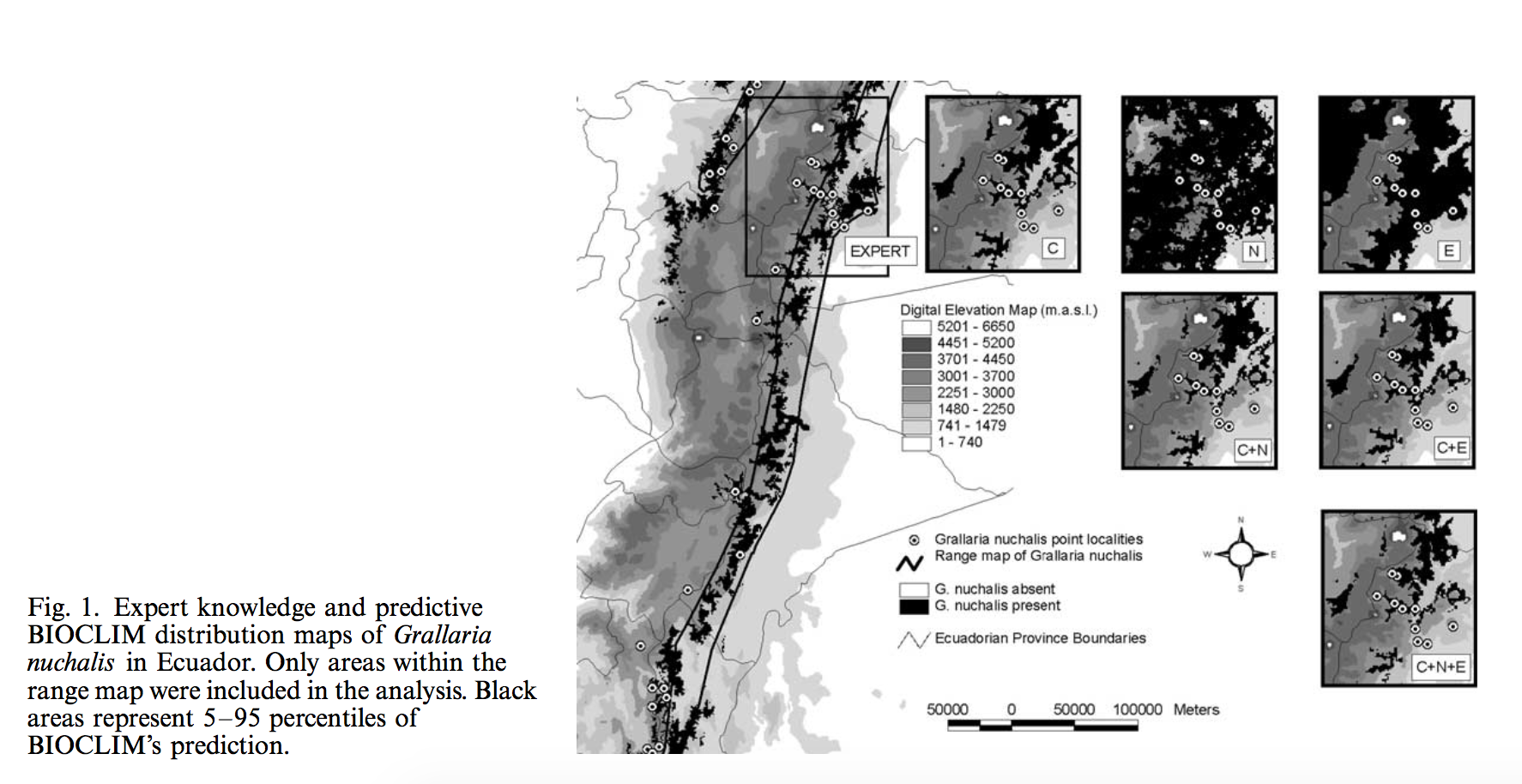
Typically, researchers use interpolated climate data or remotely sensed environmental data to build Ecological niche models (ENMs). Parra et. al. conducted the first assessment of the relative performance of models created by three different datasets: climate data, Normalized Difference Vegetation Index (NDVI), and elevation data. They compared predicted versus expected distribution of six bird species in the Ecuadorian Andes. They developed seven models based on three datasets and all their combinations using BIOCLIM. Predictive maps were compared with expert knowledge based maps, and sensitivity, specificity, positive predictive power, and Kappa were calculated. They found that models included climate variables performed well across most measures, whereas ones only use NDVI performed the worst. In the mean while, elevation data based models showed high over-prediction errors. They concluded that it is usually beneficial to include various datasets into ENMs when possible. Data quality of remote sensing data should be evaluated carefully before being included, especially for regions with complex topography or cloudy weather. This comparison result, however, may revealed a regional trend for Ecuadorian Andes but not a general rule, considering the special landscape, high levels of endemism, and species richness of the study area. Therefore, similar modeling comparison will benefit further understanding for effects of data choosing on ENMs.