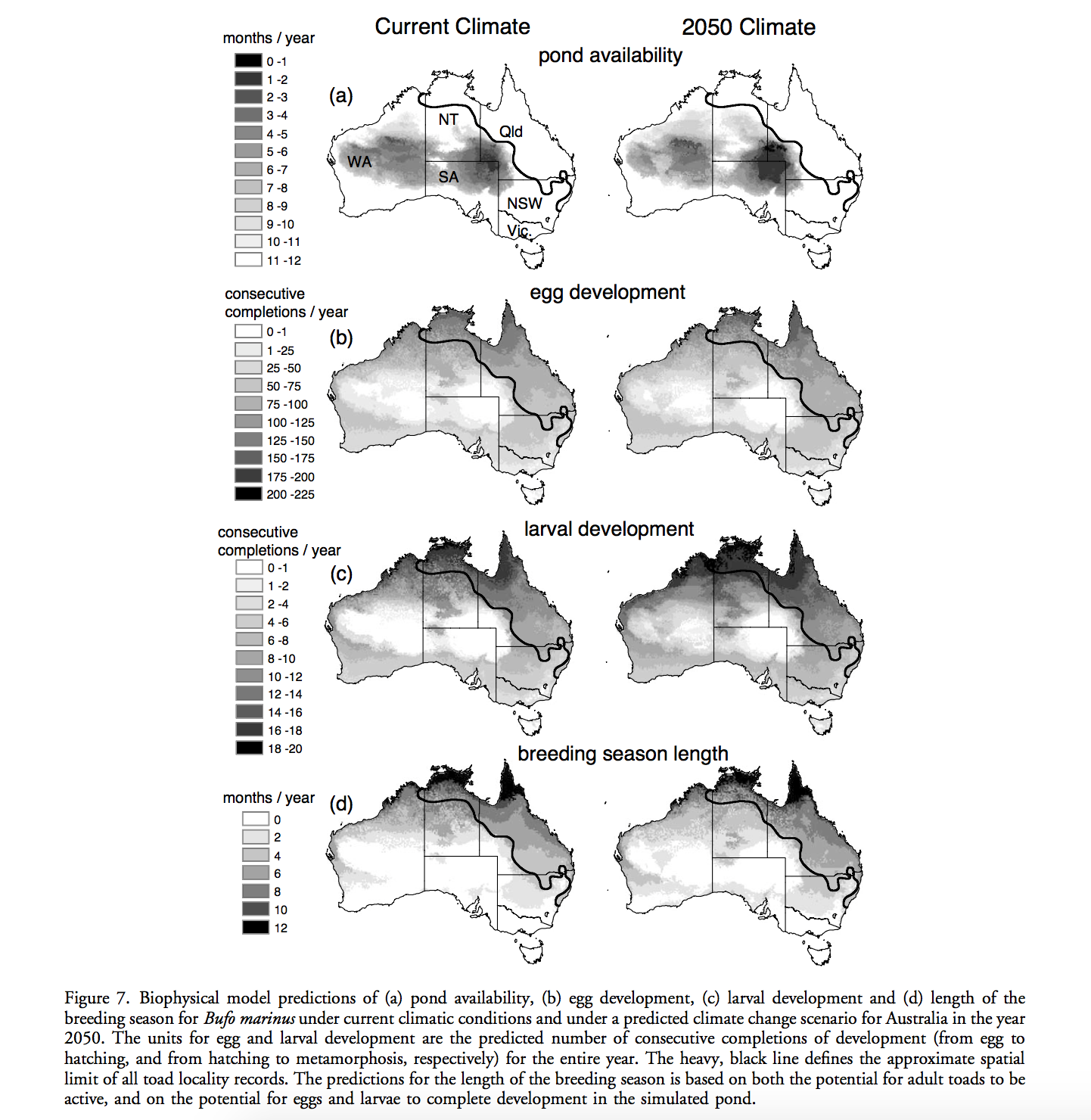
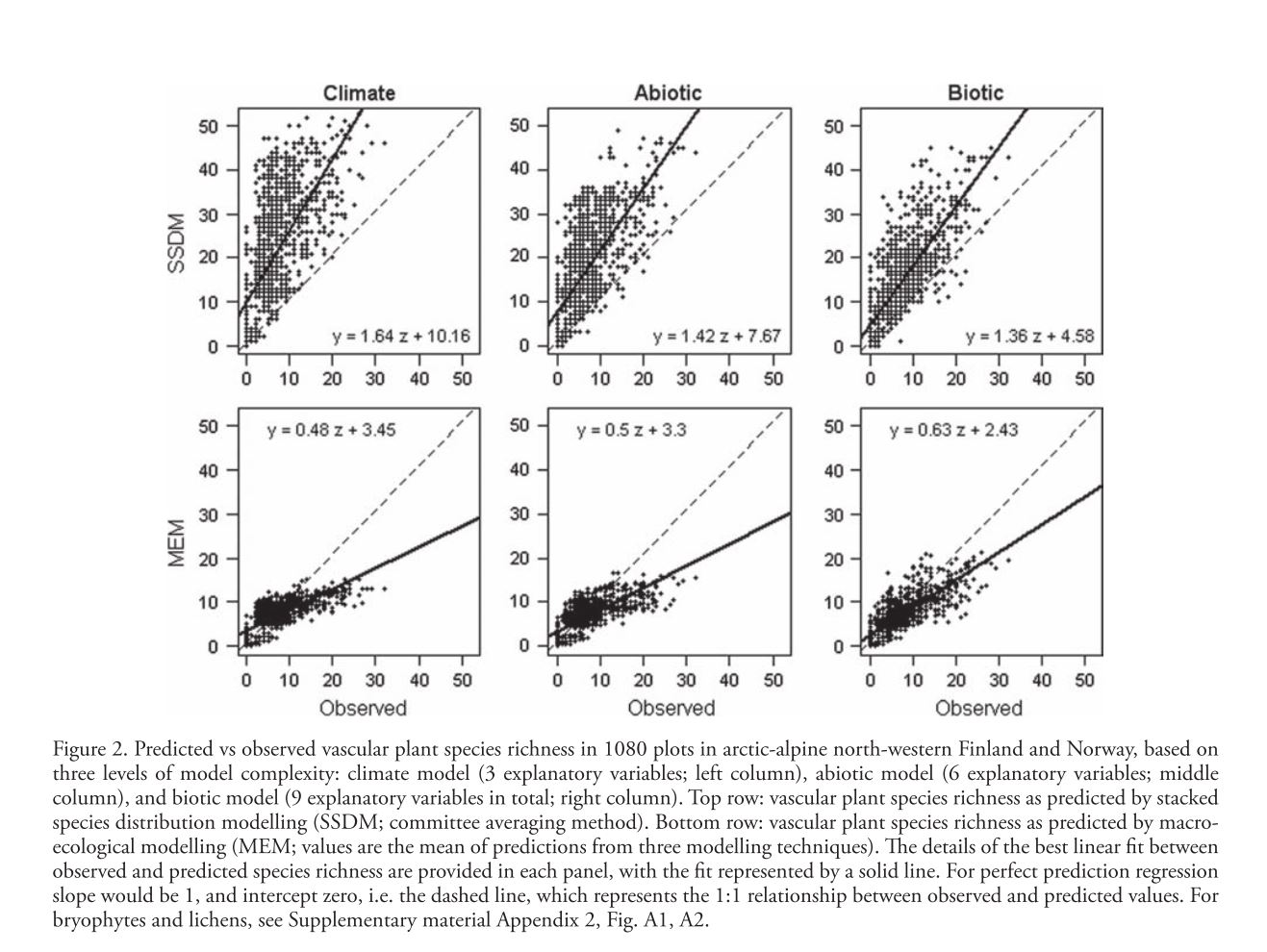
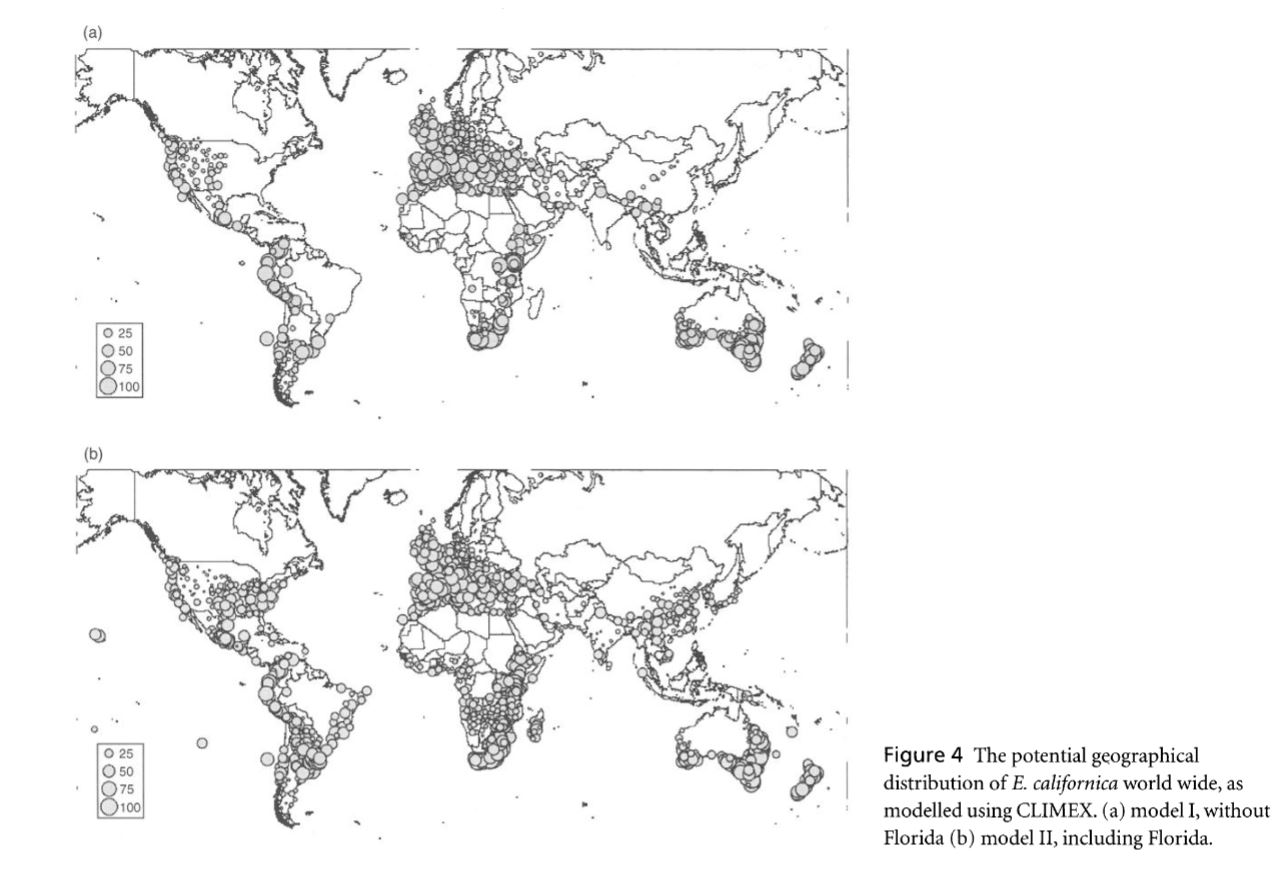
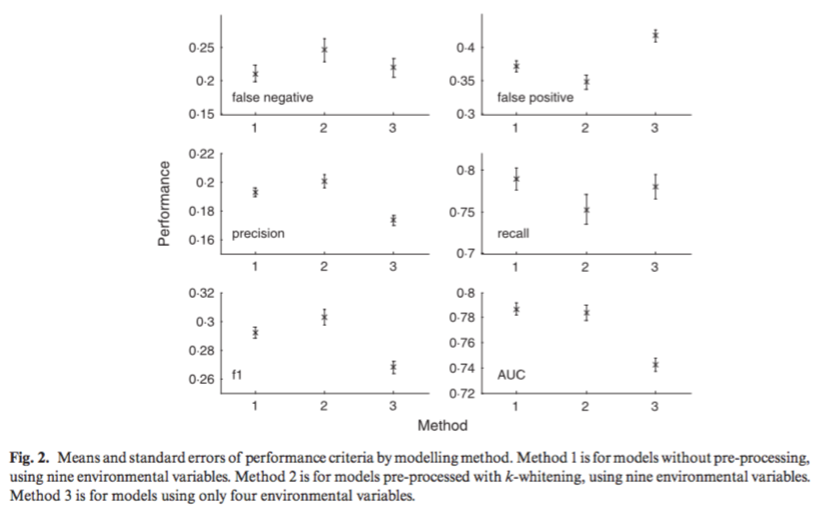
Rougier, T., Lassalle, G., Drouineau, H., Dumoulin, N., Faure, T., Deffuant, G., Rochard, E. and Lambert, P. 2015. The Combined Use of Correlative and Mechanistic Species Distribution Models Benefits Low Conservation Status Species. Plos One, 10, 21. DOI: 10.1371/journal.pone.0139194
The spatial distribution of species suitable habitat has typically been projected using correlative Species Distribution Models (SDMs). Now increasing evidence suggests that rapid evolutionary change, dispersal, spatial structure of the environment, and population dynamics are also important for determining future species ranges. This paper seeks to develop a framework for joint analysis of correlative and mechanistic SDMs in order to increase the robustness of model-derived conclusions and aid resource managers involved in species conservation planning. Two previously constructed models, one correlative and one mechanistic, were used along with biological data collected from the literature EuroDiad 3.2 database which contains distribution information for European diadromous fishes from 1750-2010. Due to computational constraints a subset (73 of the 197 river basins included in the database) were used to predict species distribution using the mechanistic model. Both the correlative and predictive model correctly predicted historical presence data (before 1900) for the river basins used, though they did not predict absences within the historical data set well. In this case both models predict a high probability of self-sustaining populations of allis shad under moderate and pessimistic climate change models. This study concludes that when available predications from correlative and mechanistic modelling should be utilized in a complementary way to help guide conservation efforts in light of climate change. While the paper does provide a framework for jointly analyzing a correlative and mechanistic distribution model it only briefly addresses the issues encountered regarding the increased intensity in the amount of data and computational power required to utilize the mechanistic SDM. For now, it may be a rare case when correlative and mechanistic SDMs can be used in a complementary way as presented in this paper.