Olden, Julian D., Joshua J. Lawler, and N. LeRoy Poff. “Machine learning methods without tears: a primer for ecologists.” The Quarterly review of biology 83.2 (2008): 171-193.
Overview
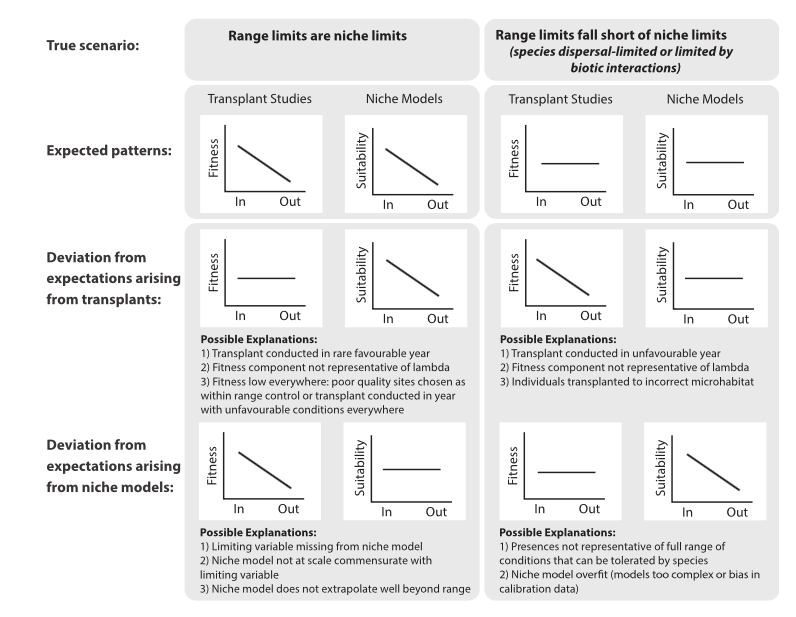
This paper was designed to help facilitate the application of machine learning techniques into the field of ecology. Authors posit that despite the advantages of machine learning techniques, ecologists have been slow in integrating these methods into their research due to lack of familiarity. It is the intent of this paper to provide background information on three categories of machine learning and examples of use in the ecological literature.
Classification and Regression Trees (Cart)
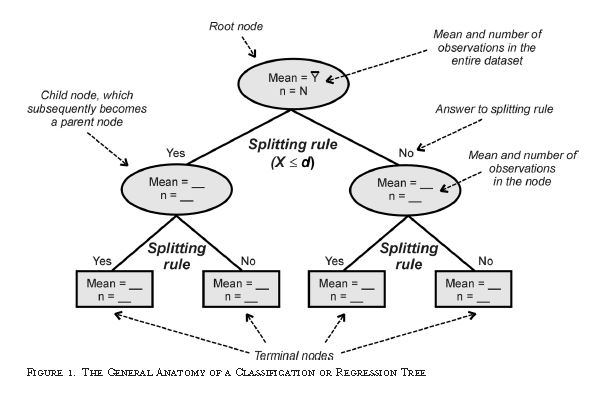
Carts can are a form of binary recursive partitioning where classification is based upon making splits within the provided dataset. The three basics of a cart algorithm involve tree building, stopping the tree building, and tree pruning and optimal selection. One of the biggest strengths of carts compared to other machine learning methods is that they’re relatively easy to interpret. One weakness is that decision trees are typically unstable.
Artificial Neural Networks
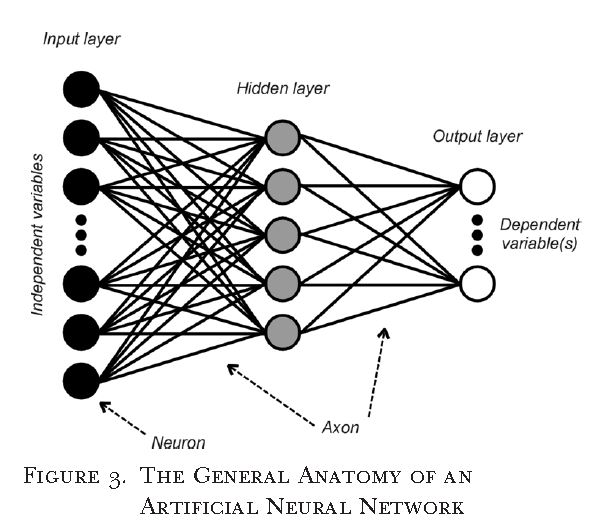
Also known as multilayer perceptrons, artificial neural networks are considered to be the universal approximations of any continuous function. Neural networks are comprised of three primary components, input layer (independent variables), While there are different types of neural networks, this paper presented the feed forward network. In a feed forward network each neuron of the previous(layer) is connected to all neurons of the next layer via axons. Connections are weighted by an activation energy function. A core strength of neural networks is that many of the underlying features can be modified to suite the research task at hand. While a weakness to neural networks would be that model performance can be sensitive to random initial conditions (weights).
Genetic Algorithm or Genetic Programming
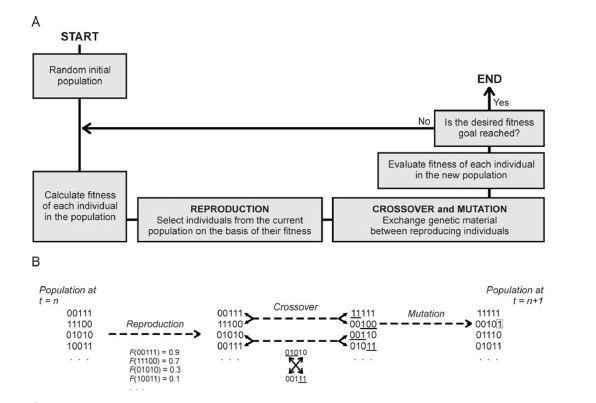
The conceptual idea behind genetic programing is that algorithms operate in a population of competing solutions to one problem, and the best solution evolves over time. Solutions are comprised of chromosomes and the chromosomes are comprised of “genes”. Genetic algorithms are strong in handling stochastic optimization procedures, but are more susceptible to model overfitting.
Conclusion
The purpose of this paper was to provide ecologist and biologist with a basic introduction to machine learning techniques and how they can be applied to ecological problems. Authors stress that while machine learning techniques can be a major boon in ecological research, machine learning will not be a end all solution to problems facing ecology. Rather, machine learning approaches should be considered an alternative to problems in massaging messy data into classical statistical models.

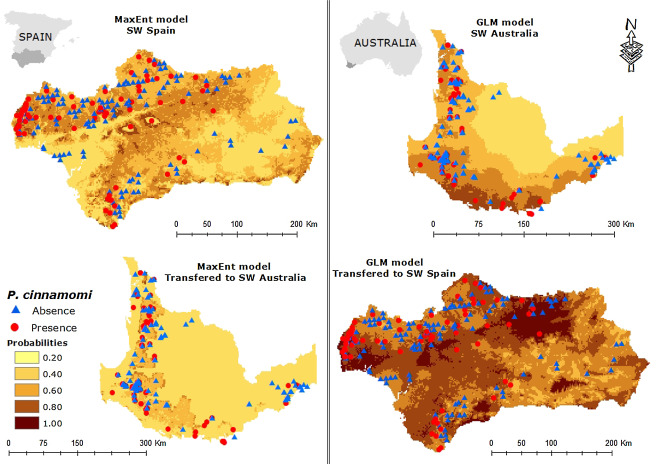 Duque-Lazo, J., van Gils, H., Groen, T. A., & Navarro-Cerrillo, R. M. (2016). Transferability of species distribution models: The case of Phytophthora cinnamomi in Southwest Spain and Southwest Australia. Ecological Modelling, 320, 62-70.
Duque-Lazo, J., van Gils, H., Groen, T. A., & Navarro-Cerrillo, R. M. (2016). Transferability of species distribution models: The case of Phytophthora cinnamomi in Southwest Spain and Southwest Australia. Ecological Modelling, 320, 62-70.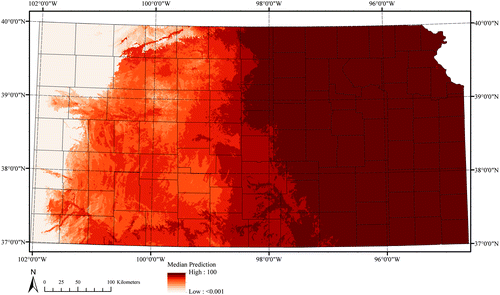
 Female adult lone star tick (Amblyomma americanum)
Female adult lone star tick (Amblyomma americanum)