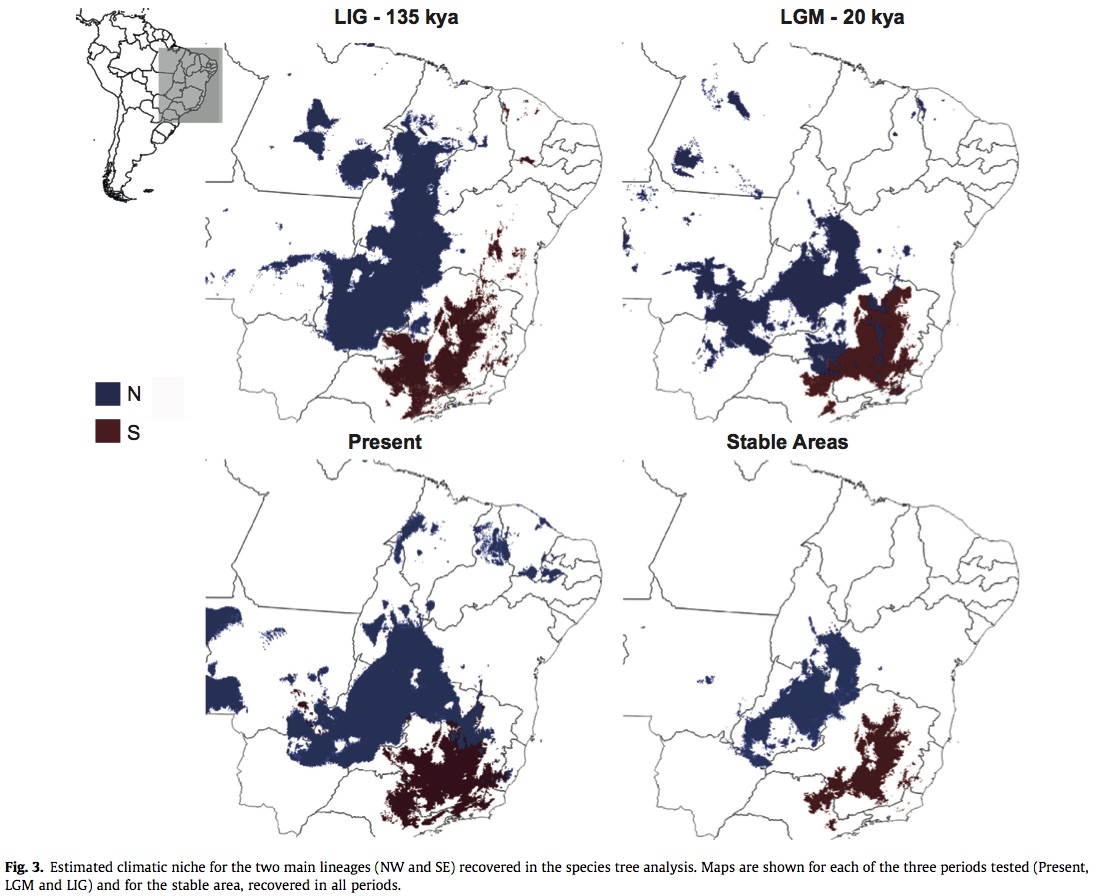
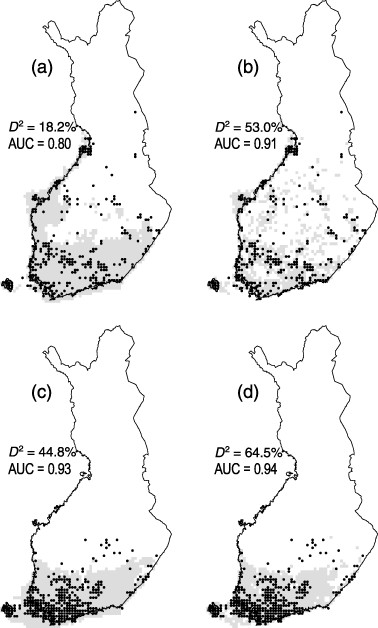
Golding and Purse suggest that Gaussian process (GP) species distribution models (SDM) via Bayesian priors may be beneficial for ecologists that wish to incorporate prior knowledge of their system and retain the speed and accuracy of predictions granted by other models. Gaussian processes are able to fit complex (i.e. more statistical terms) statistical models, but typically require computationally extensive methods (e.g. Markov chain Monte Carlo methods). Consequently, the authors evaluate another method of incorporating GP SDMs by comparing its predictive ability and run time with other commonly used approaches in a dataset from the North American Breeding Bird Survey for both presence/absence and presence-only data. Models compared in their study include: a GP model, a generalized additive model (GAM), and a boosted regression tree model (BRT). Instead of fitting GP SDM models with MCMC, they evaluate the efficacy of a more efficient deterministic inference procedure called Laplace approximation and expectation propagation. Deterministic approximations are subject to error that may decrease accuracy of predictions, but the authors argue that even with these limitations GP models fitted with deterministic inference are a promising method for SDM analyses. They found that the predictive accuracy of GP SDMs fitted by Laplace approximation was higher than BRT, GAMs, and logistic regression for presence/absence data and higher than all compared models for presence-only data. Additionally, GP SDMs were just as fast as GAMs. For situations when data on species occurrence is sparse, such vector abundance and distribution, but distributions of hosts is better documented (e.g. cattle or humans) this method would allow integration of multiple types of prior information.
Golding, N. & Purse, B.V., 2016. Fast and flexible Bayesian species distribution modelling using Gaussian processes. Methods in Ecology and Evolution. doi: 10.1111/2041-210X.